Decoding cancer Risk with AI: A Deep Dive into Predictive Modeling and Biological Interpretation
Predicting patient outcomes in cancer treatment is a complex challenge. Recent advancements in artificial intelligence, coupled with high-resolution digital pathology, are offering powerful new tools to address this need. This article details the methodology behind our research, outlining how we leveraged deep learning to assess cancer risk and, crucially, why the model makes the predictions it does – bridging the gap between algorithmic output and biological reality.you’ll gain a clear understanding of our approach, from data processing to statistical validation, and how we’re uncovering insights into the tumor microenvironment.
Data & Model Development: building a Foundation for Accurate Prediction
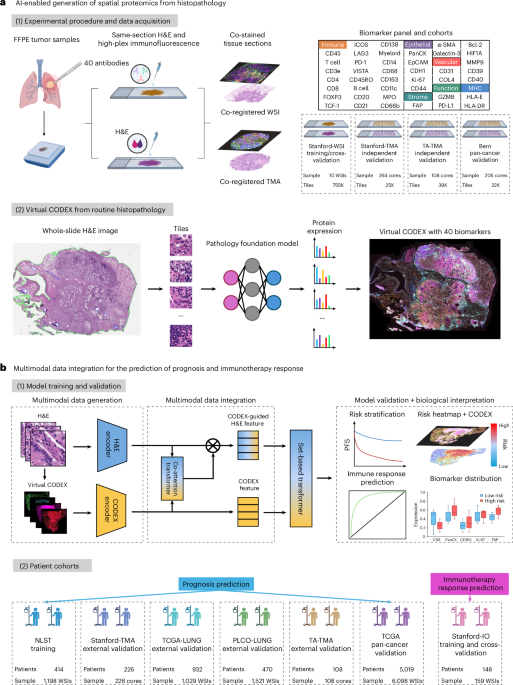
Our work centers around analyzing whole-slide images (WSIs) of Hematoxylin and Eosin (H&E) stained tissue, alongside spatially resolved proteomic data obtained through CODEX imaging. Here’s a breakdown of the key steps:
* WSI Preprocessing: We meticulously processed WSIs, ensuring consistent quality and standardization for optimal model performance. This included tile extraction – dividing the large images into smaller, manageable segments.
* Deep Learning Model: We employed a convolutional neural network (CNN) architecture, specifically a ResNet50 model pre-trained on ImageNet, to extract meaningful features from the H&E tiles. This pre-training provides a strong starting point, allowing the model to learn more efficiently from our cancer data.
* Protein Expression Prediction: The CNN’s learned features were than used to predict the expression levels of various proteins identified through CODEX imaging. This allows us to link visual patterns in H&E images to underlying biological processes.
* Risk Stratification: We evaluated the performance of our model in predicting patient outcomes using the C-index, a standard metric for assessing the discriminatory power of risk prediction models.Kaplan-Meier analysis further validated the model’s ability to stratify patients based on risk.
Unlocking the “Why” Behind Predictions: Biological Interpretation with Integrated Gradients
A powerful AI model is onyl truly valuable if we understand why it’s making certain predictions. We didn’t want a “black box”; we wanted biological insight. To achieve this,we utilized a technique called Integrated Gradients.
* Integrated Gradients Explained: This method quantifies the contribution of each input feature (in our case, each pixel within an H&E tile) to the model’s final risk prediction. positive attributions highlight areas associated with increased risk,while negative attributions indicate protective features.
* Captum Library: We implemented Integrated gradients using the Captum library (version 0.4.0), a trusted resource for explainable AI.A zero vector served as our baseline for comparison.
* Normalization & Aggregation: Integrated gradient values were normalized across each WSI to allow for meaningful comparisons.We then aggregated these scores across the entire dataset, identifying tiles with the highest and lowest risk attributions (top and bottom 1%).
* Linking to Protein Expression: For these high- and low-risk tiles, we analyzed the corresponding CODEX data to determine the average expression levels of key biomarkers. This revealed distinct protein expression profiles associated with predicted risk.
Delving into the Tumor Microenvironment: Co-Expression Analysis & cell State Characterization
To further refine our understanding, we investigated how biomarkers co-express within the tumor microenvironment. This provides clues about the functional interactions driving cancer progression.
* Biomarker Co-Expression: We identified tiles exhibiting high biomarker expression (above the 80th percentile) and assessed the frequency of co-expression between biomarker pairs.
* Spatial Cell State Analysis: We focused on six pre-defined combinations of lineage and functional markers to characterize specific cell states relevant to immunotherapy response:
* Granzyme B+/CD8+ (Cytotoxic T cells)
* TCF-1+/CD4+ (Stem-like CD4+ T cells)
* PD-1+/CD8+ (Exhausted T cells)
* CD66b+/MMP9+ (Neutrophils)
* FAP+/collagen IV+ (Cancer-associated fibroblasts)
* CD163+/MMP9+ (Tumor-associated macrophages)
*