Joseph Donovan
2026-01-15 00:00:00
The trial methodology, conduct and analysis are described in the published protocol27 and statistical analysis plan28. The trial was designed and delivered by the investigators, supported by the Oxford University Clinical Research Unit (OUCRU) Clinical Trials Unit. All authors vouch for the data and analysis. The trial was registered at ClinicalTrials.gov (NCT03100786).
Settings and study population
We recruited participants from the Hospital for Tropical Diseases and Pham Ngoc Thach Hospital for Tuberculosis and Lung Disease, in Ho Chi Minh City, Vietnam.
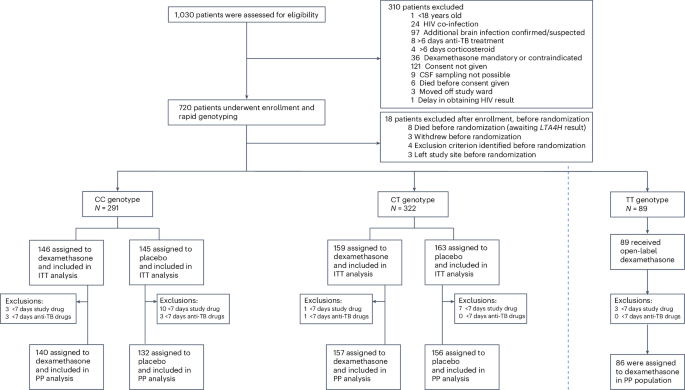
Individuals were considered eligible for enrollment if all inclusion criteria were met. As detailed in the published protocol27 inclusion criteria for the LAST ACT trial were:
-
(1)
≥18 years old
-
(2)
HIV negative
-
(3)
A clinical diagnosis of tuberculous meningitis ( ≥5 days of meningitis symptoms and consistent CSF abnormalities) with antituberculosis chemotherapy either planned or started by the treating clinician
Individuals were considered ineligible for enrollment if any exclusion criteria were met. As detailed in the published protocol27 exclusion criteria for the LAST ACT trial were:
-
(1)
An additional brain infection (other than tuberculous meningitis) confirmed or suspected: positive CSF Gram or India Ink stain, positive blood or CSF Cryptococcal antigen test and cerebral toxoplasmosis suspected and attending clinician plans to give antitoxoplasmosis treatment with antituberculosis treatment.
-
(2)
More than six consecutive days of two or more drugs active against M. tuberculosis immediately before screening.
-
(3)
More than six consecutive days of any type of orally or intravenously administered corticosteroid immediately before randomization.
-
(4)
Systemic corticosteroids were considered mandatory for any reason by the attending clinician.
-
(5)
Systemic corticosteroids were considered contraindicated for any reason by the attending clinician.
-
(6)
Patient previously been randomized into the LAST ACT trial for a prior episode of tuberculous meningitis.
-
(7)
Lack of consent from the participant or family member (if the participant is incapacitated by the disease).
Participants were subsequently classified as having definite, probable, or possible tuberculous meningitis, following published diagnostic criteria26 (Supplementary Table 55).
Study oversight
Written informed consent to enter the trial was obtained from all participants or a relative if they were incapacitated. If capacity returned, consent from the participant was obtained. Trial approvals were obtained from local and national ethics and regulatory authorities in Vietnam, and the Oxford Tropical Research Ethics Committee in the UK (Supplementary Text 1). An independent data monitoring committee (Supplementary Text 2) reviewed data, following a prespecified plan, at 6-monthly intervals for the first 24 months of enrollment and annually thereafter.
LTA4H genotyping
Once a participant or their relative consented to enter the study, blood was taken for rapid LTA4H genotyping. Genomic DNA was extracted from 5 ml of venous blood collected in tubes containing EDTA anti-coagulant using the Nucleon BACC2 Genomic DNA extraction kits (GE Healthcare). DNA quality and concentration were determined by Nanodrop 2000 v1.0 Spectrophotometer (Thermo Scientific). rs17525495 was genotyped by Taqman using predesigned assay kits (Applied Biosystems) according to the supplier’s instructions (https://documents.thermofisher.com/TFS-Assets/LSG/manuals/MAN0009593_TaqManSNP_UG.pdf). We performed real-time PCR with 15 ng of DNA using a LightCycler 480 Probes Master kit on the LightCycler 480 real-time PCR system (Roche) according to the supplier’s protocol (https://pim-eservices.roche.com/LifeScience/Document/4dd0e207-97ed-e311-98a1-00215a9b0ba8). For each PCR run, the DNA samples from each participant were analyzed in duplicate, and three positive control samples for TT, CC homozygous and CT heterozygous were included. These control samples were from samples that had been previously genotyped for rs17525495 by Taqman, as reported elsewhere29 and confirmed by Sanger sequencing. The genotyping results were then analyzed using LightCycler480 software.
Randomization and study groups
Randomization occurred once LTA4H genotyping results were available, usually within 24 h. LTA4H CC- and CT-genotype participants were randomized to two parallel groups in a 1:1 ratio: dexamethasone or placebo for 6–8 weeks. TT-genotype participants received open-label dexamethasone for 6–8 weeks.
Randomization was stratified by participating hospital, LTA4H genotype and modified MRC disease severity grade30 assessed at enrollment. Participants in grade I had a Glasgow coma score of 15 (possible range 3–15, with higher scores indicating better status) with no focal neurologic signs, grade II participants had a score of either 11–14 or had focal neurological signs and grade III participants had a score of 10 or less. The randomization list was computer-generated based on random permuted blocks with block size 4 and 6 (probability 0.75 and 0.25). Participant randomization was performed by trained clinical staff using a web-based software, with 24-h availability.
Study treatments
All participants received standard-of-care antituberculosis chemotherapy according to national guidelines. Rifampicin (10 mg kg−1 per 24 h, maximum 600 mg), isoniazid (5 mg kg−1 per 24 h, maximum 300 mg), pyrazinamide (25 mg kg−1 24 h, maximum 2 g) and ethambutol (20 mg kg−1 per 24 h, maximum 1.2 g) were given for at least the first 2 months of treatment, provided drug resistance was not suspected or proven. Pyrazinamide was then stopped and rifampicin, isoniazid and ethambutol (at the same doses) were given until at least 12 months antituberculosis treatment in total had been given. Patients with visual complications discontinued ethambutol and an alternative drug was used in its place. The decision of which alternative fourth drug to use is made by the treating physician and was not defined by the trial, but in practice was either levofloxacin or streptomycin or amikacin.
For tuberculous meningitis caused by isoniazid-resistant tuberculosis the attending physician decided which drugs to prescribe, dependent upon clinical circumstances, but usually isoniazid was substituted by levofloxacin. For participants with multidrug resistant tuberculosis, second-line treatment was given as soon as possible, following national guidelines and local policies.
Participants allocated dexamethasone received the 6–8-week regimen previously shown to reduce tuberculous meningitis mortality8. Patients with grade II or III disease received intravenous treatment for four weeks (0.4 mg per kilogram per day for week 1, 0.3 mg per kilogram per day for week 2, 0.2 mg per kilogram per day for week 3 and 0.1 mg per kilogram per day for week 4) and then oral treatment for 4 weeks, starting at a total of 4 mg per day and decreasing by 1 mg each week. Patients with grade I disease received three weeks of intravenous therapy (0.3 mg per kilogram per day for week 1, 0.2 mg per kilogram per day for week 2 and 0.1 mg per kilogram per day for week 3) and then 3 weeks of oral therapy, starting at a total of 3 mg per day and decreasing by 1 mg each week. We followed this drug dosage regimen, as it was the one which proved beneficial to adults in our previous trial8. No other corticosteroid regimens, including oral regimens, are supported by such strong trial data; therefore, we could not justify using alternative drugs or doses. For our current trial, patients with grade I disease received intravenous therapy during week 3, whereas in our previous trial8, patients with grade I disease received oral therapy during week 3 (albeit at 0.1 mg per kilogram per day in both trials). For the trial published in the year 20048, we chose to have a shorter regimen for the least severe patients (grade 1) for pragmatic reasons: at the time doctors felt those with less severe disease were usually discharged home within 3 weeks and therefore it was not acceptable to have a 4-week intravenous regimen. Individuals with more severe disease usually stayed in hospital for longer, and longer intravenous treatment was both desirable and possible. This regimen is now the global standard-of-care.
Blinded fully made-up and labeled study treatment packs contained either dexamethasone or identical placebo. All participants and investigators were blinded to study drug allocation. Adherence to medication was ensured with the use of supervised drug intake for inpatients, encouraged by detailed instructions at discharge, and medication compliance checks at follow-up visits or phone calls.
Outcome assessments
The primary endpoint was all-cause mortality or new neurological event during 12 months from randomization. A new neurological event was defined as a Glasgow coma score reduction by ≥2 points for ≥2 days from the highest previous score or the new onset of cerebellar symptoms, focal neurological signs or seizures.
Secondary endpoints, assessed over 12 months from randomization, were overall mortality, first new neurological event, neurological disability (modified Rankin score 3–5) (Supplementary Table 56), modified Rankin score as an ordinal scale, use of open-label corticosteroid treatment for any reason, serious or severe adverse events, and resolution of CSF and blood inflammation. The Rankin scale assesses dependence. A score of 0 indicated no symptoms; 1 indicated minor symptoms not interfering with lifestyle; 2 indicated symptoms that might restrict lifestyle, but patients could look after themselves; 3 indicated symptoms that restricted lifestyle and prevented independent living; 4 indicated symptoms that prevented independent living, although constant care and attention were not required; and 5 indicated total dependence on others, requiring help day and night.
Clinical assessments
Participants underwent clinical assessments at baseline—days 3, 7, 10, 14, 21 and 30—and monthly until month 12. Assessment included Glasgow coma score, focal neurological deficits and details of adverse events. Participants were monitored daily whilst in hospital and serious adverse events were reported to local and national regulators. In participants for whom systemic corticosteroids were considered necessary by the treating clinician after randomization, study drug was discontinued (with previous doses remaining blinded) and corticosteroids commenced.
CSF and whole blood analyses
Lumbar CSF was sampled at baseline and on days 30 and 60. At least 6 ml of CSF (if available) was used for Ziehl–Neelsen smear microscopy, either Xpert MTB/RIF or Xpert MTB/RIF Ultra, and mycobacterial culture (mycobacteria growth indicator tube) following standard procedures31. Phenotypic drug susceptibility testing was performed using a BACTEC mycobacteria growth indicator tube SIRE kit (Becton, Dickinson).
Preplanned analyses of CSF proteomic and whole blood transcriptomic data were performed, focused on six targeted pathways known to be important mediators of tuberculous meningitis (TBM) pathogenesis: CSF IFN signaling, neutrophil degranulation, neutrophil activation, eicosanoid metabolic processes, TNF signaling, and cytokine signaling, and ten reported cytokines. CSF inflammatory proteins were measured using the Olink Explore 384 Inflammation Panel (Olink Proteomics). Olink measurements were conducted for 675 participants on day 0 (n = 675) and day 30 (n = 397) at the Human Genomics Facility of the Genetic Laboratory, Department of Internal Medicine, Erasmus MC. Raw protein expression data were reported as normalized protein expression units, which were log2-transformed and normalized using the plate control method to minimize technical variation. To correct for batch effects, the normalized protein expression values then were further adjusted using the ComBat function from the sva R package32. Quality control procedures were conducted at both the sample and protein levels. At the sample level, poor-quality samples were excluded if they exhibited a failure rate of ≥50% across protein assays, as determined by Olink’s internal quality control criteria. Outliers were identified via principal component analysis and excluded if they deviated by more than three standard deviations from the first principal component. At the protein level, proteins with a limit of detection exceeding 75% of samples were filtered out. Finally, 17 participants who died before randomization were excluded, resulting in a final dataset of 275 proteins in 1,029 CSF samples from 646 participants available for analysis (day 0: n = 638; day 30: n = 391). Among the ten planned CSF cytokines—TNF, IL-1β, IL-2, IL-6, IL-12β, IFN-γ, IL-4, IL-5, IL-10 and IL-13—four (IL-2, IL-4, IL-5 and IL-13) did not pass quality control due to their limit of detection exceeding 75% of samples.
Whole-blood RNA sequencing was performed for the first 207 consecutively enrolled participants on day 0 (n = 207), day 14 (n = 191) and day 60 (n = 156). Whole-blood samples were preserved in PAXgene Blood RNA collection tubes at −80 °C. Total RNA was subsequently extracted using the PAXgene Blood RNA Kit (Qiagen, Valencia), following the manufacturer’s protocol. Extracted RNA was shipped to the Ramaciotti Centre for Genomics (University of New South Wales) for high-throughput sequencing. Library preparation was performed using the TruSeq Stranded Total RNA with Ribo-Zero Globin kit (Illumina) to deplete globin transcripts and ribosomal RNA. Sequencing was conducted on the Illumina NovaSeq 6000 platform, generating approximately 30 million 100 bp paired-end reads per sample. Raw sequencing data were subjected to quality control and aligned to the human reference genome (GRCh38 build 99) with the STAR aligner (v2.5.2a)33. Gene-level quantification from aligned reads was performed using FeatureCounts (v2.0.0), generating raw counts for 60,067 genes34. Before analysis, five participants were excluded: two who died before randomization and three whose RNA sequencing data were poor quality (RNA integrity number n = 202, day 14: n = 188, day 60: n = 153). To further clean the dataset, hemoglobin genes, ribosomal RNA genes and genes with low expression (median count 2-transformed using the variance stabilizing transformation algorithm implemented in the DESeq2 package in R (v1.34.0) to enable downstream statistical analyses35. For each targeted pathway, a single sample enrichment score was calculated using the z-score method to evaluate the activity of the pathway at each timepoint for both whole blood transcriptomics and CSF proteomics, for each patient.
Statistical analyses
We adopted a hybrid trial-design approach that aimed to prove noninferiority of placebo first but also superiority of placebo should dexamethasone prove harmful. The trial had two primary populations: the combined CC- and CT-genotype population and the CC-genotype population28.
The primary analysis used a Cox proportional hazards regression model with the primary endpoint as the outcome, where we aimed to prove noninferiority of placebo in the CC- and CT-genotype population or the CC-genotype subgroup. We corrected for multiple testing using the method by Spiessens and Debois36. It takes the correlation into account between the test statistic for the CC-genotype subgroup analysis and the overall CC- and CT-genotype analysis, which makes it less conservative than the Bonferroni approach. We chose to spend 2% of the one-sided type I error of 2.5% to the CC- and CT-genotype analysis, leaving 0.86% for the CC-genotype subgroup analysis.
In addition, we evaluated the primary endpoint using a superiority design, in the CC- and CT-genotype population, in the CC-genotype subgroup and in the CT-genotype subgroup. In the superiority analyses, we did not make any correction for multiple testing.
We set the noninferiority margin in favor of dexamethasone at a hazard ratio of 0.75 and assumed a true hazard ratio of 1.15 in the CC- and CT-genotype population. To obtain 80% power at the one-sided 2% significance level, 184 events in the CC- and CT-genotype population would be required. Assuming a 12-month risk of the primary endpoint in the dexamethasone arm of 35%, a hazard ratio of 1.15 corresponds to a risk of 31.2% of placebo, and the noninferiority margin implies that we can exclude an absolute risk increase of placebo of (at worst) +8.7%. Assuming an overall event risk of ≥32% and 11% sample increase to compensate for loss-to-follow-up, we aimed to randomize 640 CC- and CT-genotype participants. Anticipating 10% being LTA4H TT genotype, we planned to enroll 720 participants in total.
The analysis followed a prespecified and published plan28. The data were analyzed using the program R (version 4.4.2; R Core Team, 2024)37. In brief, intention-to-treat and per-protocol analyses were performed for the primary and secondary endpoints. The intention-to-treat analysis included all randomized CC- and CT-genotype participants, even if no study drug was received after randomization. The per-protocol analysis included all randomized participants, excluding those subsequently found to have not met all inclusion criteria or to have met any exclusion criteria and participants with a final diagnosis other than tuberculous meningitis. We also excluded participants who received
Baseline characteristics were summarized by treatment arm and genotype for intention-to-treat and per-protocol analyses. The primary analysis was a Cox proportional hazards regression model with the primary endpoint as the outcome, treatment as the only covariate and with LTA4H genotype (CC or CT) and modified MRC severity grade at enrollment as stratum variables. We also performed the analysis in CC-genotype participants only. The null hypothesis of the noninferiority comparison was tested via CIs for the hazard ratio. Further analyses used a superiority design, using hazard ratio and difference in RMTL as effect measures for the time-to-event outcomes and logistic regression and a proportional odds regression model for modified Rankin scores. For the Rankin score R = r, where 1 ≤ r ≤ 6, the proportional odds regression model quantifies the odds of having a score of r or higher for a participant allocated to dexamethasone, compared with a participant allocated to placebo, that is, (Probability(R ≥ r|Dexamethasone)/Probability(R r|Dexamethasone))/(Probability(R ≥ r|Placebo)/Probability(R r|Placebo)). In the regression equation we allow the intercept to differ by r, whereas the relation of the covariables with the odds is assumed not to depend on r.
Subgroup analysis of the primary endpoint was planned for LTA4H genotype, and enrollment modified MRC severity grade, tuberculous meningitis diagnosis (definite, probable and possible) and M. tuberculosis drug susceptibility profile. Analyses were conducted for CC and CT genotypes combined and CC genotype alone, with the exception of adverse events, which were compared by genotype. No corrections were made for multiple testing, except for the CSF inflammation and blood analysis that focused on predefined pathways and molecules of known importance to tuberculous meningitis pathogenesis38. Linear mixed-effects models and Bayesian approaches described LTA4H genotype effects on baseline transcriptomic and proteomic signatures and the longitudinal effects of treatment and genotype upon them.
We compared TT-genotype participants with dexamethasone- and placebo-treated CC- and CT-genotype participants for mortality, new neurological events, disability and CSF/blood inflammation. An individual participant data meta-analysis was planned to investigate dexamethasone’s effect on survival and disability in HIV-negative adults with TBM overall, independent of LTA4H genotype28. Current trial data were combined with 447 HIV-negative adults with tuberculous meningitis from the 2004 trial8.
Some exploratory analyses (not articulated in the statistical analysis plan) were conducted and presented. These include the effect of MRC grade on dexamethasone’s effect on survival across the three genotypes (Supplementary Fig. 6); a comparison of day 30 CSF cytokines and CSF inflammatory pathways between participants of CC and CT genotype (Supplementary Figs. 23 and 24); the association of day 30 CSF cytokines and CSF inflammatory pathways with the trial’s primary outcome (Supplementary Table 52); the effect of MRC grade on dexamethasone survival effect in the combined data from the two trials (Supplementary Fig. 25b); and a comparison of 9-month survival between the two trials (Supplementary Fig. 25c).
Protocol amendments
Major amendments made to the LAST ACT protocol after study start, relevant to these results, are:
-
(1)
A change in an exclusion criterion to allow no more than 6 days of any type of orally or intravenously administered corticosteroid immediately before enrollment (previously no more than 3 days).
-
(2)
Randomization to be stratified by tuberculous meningitis grade with grade set at enrollment, even if this grade had changed by randomization (which can only be performed once LTA4H genotyping results are available).
-
(3)
Serum potassium to be checked and recorded at baseline. Values to be recorded if they are repeated as part of routine clinical care.
-
(4)
Following discharge, participants to be followed up either in the hospital’s outpatient department or via a phone call. In-person or phone call outpatient review to occur monthly (±7 days) for at least the first 2 months following hospital discharge. If a patient is discharged at day 21 (for example, a grade 1 patient after 3 weeks of intravenous study drug), they are not required to return for follow up at day 30 (month 1). Disability questions due to be asked at day 30 will instead be asked at day 21.
These protocol amendments were approved by all necessary ethical committees
Protocol deviations
Major deviations from the LAST ACT protocol after study start, relevant to these results, are as follows: nine participants were discharged from hospital while still within the intravenous study drug period (in eight cases, this was done in accordance with the participant’s wishes). In each case, intravenous study drug was given as per schedule up to the point of early discharge. The oral study drug period was then started at discharge. The remaining days of intravenous study drug were not replaced, resulting in a shorter total duration of study drug than originally scheduled. Details of participants with shortened study drug regimens, in order of enrollment, are as follows:
-
(1)
The participant received a complete week of 0.4 mg kg−1 intravenously and 4 days of 0.3 mg kg−1 intravenously, followed by completion of the scheduled oral study drug.
-
(2)
The participant received a complete week of 0.4 mg kg−1 intravenously and 5 days of 0.3 mg kg−1 intravenously, followed by completion of the scheduled oral study drug.
-
(3)
The participant received a complete week of 0.4 mg kg−1 intravenously and 3 days of 0.3 mg kg−1 intravenously, followed by completion of the scheduled oral study drug.
-
(4)
The participant received a complete week of 0.4 mg kg−1 intravenously, a complete week of 0.3 mg kg−1 intravenously, a complete week of 0.2 mg kg−1 intravenously and 2 days of 0.1 mg kg−1 intravenously, followed by completion of the scheduled oral study drug.
-
(5)
The participant received a complete week of 0.4 mg kg−1 intravenously and 5 days of 0.3 mg kg−1 intravenously, followed by completion of the scheduled oral study drug.
-
(6)
The participant received a complete week of 0.4 mg kg−1 intravenously, a complete week of 0.3 mg kg−1 intravenously, a complete week of 0.2 mg kg−1 intravenously and 5 days of 0.1 mg kg−1 intravenously, followed by completion of the scheduled oral study drug.
-
(7)
The participant received a complete week of 0.4 mg kg−1 intravenously, a complete week of 0.3 mg kg−1 intravenously and 5 days of 0.2 mg kg−1 intravenously, followed by completion of the scheduled oral study drug.
-
(8)
The participant received a complete week of 0.4 mg kg−1 intravenously, a complete week of 0.3 mg kg−1 intravenously and 2 days of 0.2 mg kg−1 intravenously, followed by completion of the scheduled oral study drug.
-
(9)
The participant received a complete week of 0.4 mg kg−1 intravenously and 5 days of 0.3 mg kg−1 intravenously, followed by completion of the scheduled oral study drug.
Ethics and inclusion statement
Our trial was designed and conducted by researchers and clinicians, all of whom were living and working within Vietnam. We included local researchers and clinicians in the study design, study implementation and authorship of this publication. Local researchers gave input into study protocol design including the schedule of study investigations and performed recruitment of trial participants routine clinical care and study procedures. Contributions of local researchers to authorship are detailed in the ‘Author contributions’ section and include: data curation, formal analysis, investigation, methodology, resources, software, supervision and visualization. The research is locally relevant and was determined in collaboration with local partners. Tuberculous meningitis is the most severe form of tuberculosis and is regularly encountered in the study setting. Interventions that could optimize therapy and potentially reduce poor clinical outcomes in this disease are locally relevant and would benefit the communities involved in this study. Roles and responsibilities were agreed amongst collaborators ahead of the research, and capacity-building plans for local researchers were discussed. Local clinical, laboratory and researcher staff were trained in the conduct of high quality clinical trial research, building capacity for future studies at these sites. Trial approvals were obtained from local and national ethics and regulatory authorities in Vietnam, and the Oxford Tropical Research Ethics Committee in the UK. Ethical approvals were as follows: The Oxford Tropical Research Ethics Committee (approval no. 52-16), The Ethical Committee of the Hospital for Tropical Diseases (approval no. 37/HDDD), The Ethical Committee of Pham Ngoc Thach Hospital for Tuberculosis and Lung Disease (approval no. 1034/HDDD-PNT) and The Vietnam Ministry of Health (approval no. 151/CN-BDGDD). Local and regional research relevant to this study has been considered in citations. In line with a reimbursement standard operating procedure at OUCRU, participants (and one relative to attend follow-up visits) received reimbursement for travel expenses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.