Rigorous Trial Design & Statistical Approach for Evaluating Novel Targeted Therapy
This clinical trial was meticulously designed to assess the efficacy adn safety of a novel targeted therapy (TT) compared to standard of care (SoC) across a diverse range of advanced malignancies. A robust statistical plan, proactive monitoring, and transparent reporting were central to ensuring the reliability and validity of the findings. This document details the key elements of the study’s sample size justification, statistical analysis plan, and reporting procedures, demonstrating a commitment to scientific rigor and data integrity.
Sample Size Determination: Powering for Meaningful Clinical Difference
The primary objective of this study was to demonstrate a clinically meaningful improvement in objective response rate (ORR) with TT compared to SoC. Based on preliminary data and established benchmarks, we hypothesized that TT would achieve a 20% ORR, while SoC would yield a 5% ORR. To detect a 15% absolute difference between the treatment arms with sufficient statistical power, a formal sample size calculation was performed.
This calculation considered a one-sided chi-square test, an alpha level of 0.10 (allowing for a controlled risk of false positive findings), and a beta level of 0.20, equating to 80% power to detect a true difference if it exists. The initial calculation indicated a requirement of 86 patients per treatment arm across four pre-defined strata, totaling 344 patients.
These strata were defined based on the site of primary tumor to ensure balanced representation of key cancer types:
* Stratum A: Breast Cancer
* Stratum B: Non-Colorectal Gastrointestinal Cancers
* Stratum C: Non-Small Cell Lung Cancer (NSCLC)
* Stratum D: Other Malignancies
Competitive enrollment was implemented across these strata to facilitate timely completion of the study. The overall ORR was planned to be assessed using a two-sided Cochran-Mantel-Haenszel (CMH) test, a conservative approach to account for potential confounding variables. An unstratified sensitivity analysis was also planned to validate the findings.
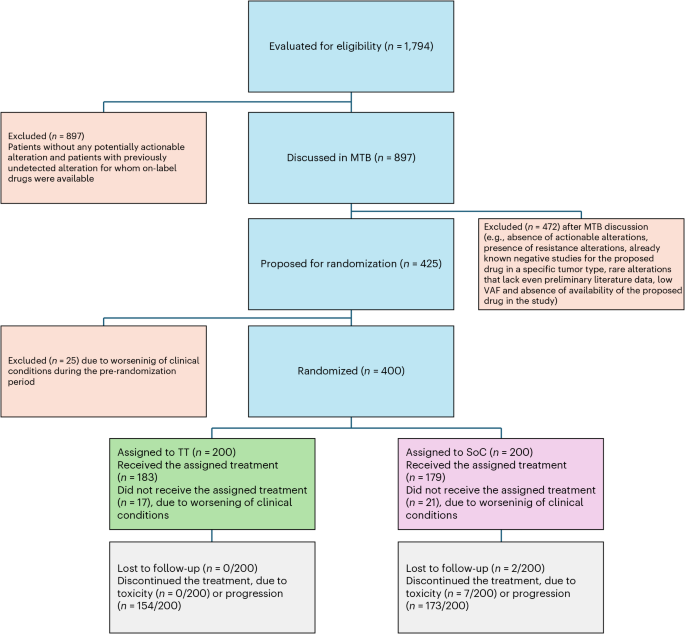
Recognizing the potential for patient dropout during the study, a 10% increase was applied, bringing the target enrollment to 384 patients. Moreover, anticipating that approximately 30% of screened patients would harbor actionable genomic alterations qualifying them for the trial, an initial screening target of 1,280 patients was established.
Addressing Screening Challenges & Final Enrollment
During the screening process, the rate of screening failures – primarily due to the complexity of genomic testing and unexpected patient ineligibility based on mutational profiles – proved higher than initially projected. The extended timelines required for comprehensive genomic assessment and the inherent uncertainty in identifying eligible patients presented logistical challenges.
to mitigate these challenges and ensure the study could be completed within a reasonable timeframe, the steering committee approved the enrollment of an additional 12 patients who were undergoing screening at the time the target of 384 randomized participants was reached. This resulted in a final enrolled cohort of 400 patients, ensuring sufficient statistical power while acknowledging the real-world complexities of genomic-driven clinical trials.
Statistical Analysis Plan: A Comprehensive & Transparent Approach
All statistical analyses were conducted using SAS software (version 9.4), a widely recognized and validated platform for clinical trial data analysis. A detailed Statistical Analysis Plan (SAP – version 1.0) was finalized prior to database lock, ensuring that all analyses were pre-specified and minimizing the risk of data-driven bias. The randomization list was generated independently by a qualified statistician using the PROC PLAN procedure within SAS and seamlessly integrated into the electronic Case Report Form (eCRF) system for automated treatment assignment.
A soft lock of the database occurred on July 31, 2024, followed by a thorough data review meeting. This led to the finalization of SAP version 2.0 on January 30, 2025, which governs all analyses presented in this report. The complete SAP document is available upon reasonable request, demonstrating our commitment to transparency and reproducibility.
Key Analytical Methods Employed:
* Descriptive Statistics: Continuous variables were summarized using mean, standard deviation (s.d.), and quartiles. categorical variables were presented as frequency distributions.
* Comparative Analyses: differences in baseline clinical characteristics between treatment arms were evaluated using appropriate statistical tests, including t-tests, binomial tests, chi-squared tests, and the CMH test.
* Homogeneity Testing: The Breslow-Day test was utilized to assess the consistency of odds ratios across different subgroups.
* **Safety