Decoding Tumor Evolution: A Deep Dive into Neoantigen Selection and Analysis in Cancer Immunotherapy
Understanding the unique fingerprint of your cancer – its mutations – is crucial for developing effective immunotherapies. This article details the rigorous process used to identify and analyze these mutations, specifically focusing on how we pinpoint neoantigens to stimulate your immune system. We’ll break down the methods employed, from whole-exome sequencing to statistical analysis, ensuring transparency and clarity.
Identifying Somatic Mutations with Whole-Exome Sequencing (WES)
The journey begins with whole-exome sequencing (WES), a powerful technique to map the genetic landscape of both your tumor and healthy (germline) DNA. This allows us to identify somatic mutations – those arising specifically within the tumor – that differentiate it from your normal cells.
Here’s how the process unfolds:
WES is performed on both tumor and germline samples.
A validated bioinformatics tool, GEM ExTra pipeline NG2-LDT 1.14.0 (Natera), meticulously compares the sequences. This identifies single-nucleotide mutations present in the tumor but absent in your germline DNA.
Alignment to a standard reference: All data is aligned to the NCBI GRCh37 human genome assembly, ensuring consistency and comparability.
Filtering for impactful mutations: To focus on the most relevant changes, stop-gain and start-loss mutations – those that directly disrupt protein production – were excluded from further analysis.
Selecting Neoantigens for Targeted Immunotherapy
Once somatic mutations are identified, the next step is to pinpoint neoantigens – mutated proteins that your immune system can recognize as foreign.This is where precision is paramount. Up to ten neoantigens per patient are selected from the list of somatic single-nucleotide variations (SNVs) generated by WES.
Random selection, not algorithms: Importantly, these neoantigens aren’t chosen by an algorithm. Rather, thay are randomly selected, allowing for a broader depiction of potential immune targets. Peptide Design: An 18-mer sequence, generally centering on the mutation, is designed.
Synthesis of Overlapping Peptides: Genscript then synthesizes two 15-mer peptides that overlap by 11 amino acids,fully covering the 18-mer sequence. These peptides are the building blocks for testing your immune response. (Detailed sequences can be found in Supplementary Table 1).
Rigorous Statistical Analysis for Meaningful Insights
Analyzing the data generated from neoantigen testing requires robust statistical methods. We employ a multi-faceted approach to ensure the reliability and clinical relevance of our findings.
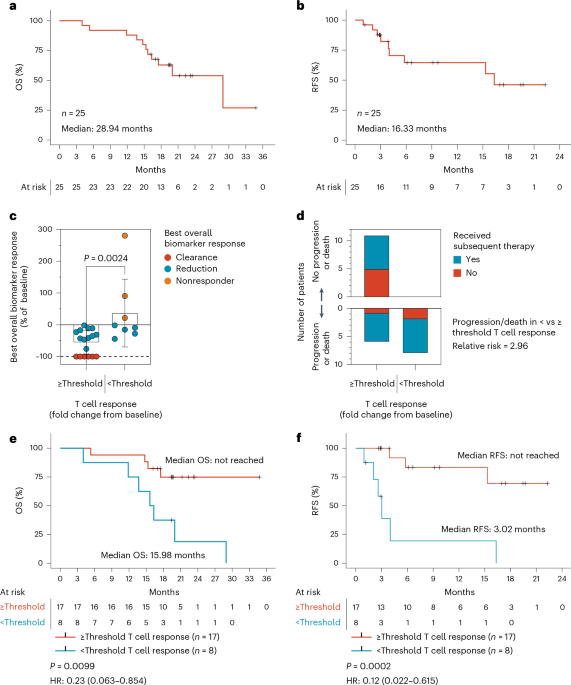
Descriptive Statistics: We summarize patient demographics, medical history, and safety data using means, standard deviations, medians, and ranges for continuous variables. Frequency counts and percentages are used for categorical variables. Efficacy Assessment: Clinical efficacy,such as tumor biomarker reduction or clearance,is examined in relation to T cell response levels. The Mann-whitney test helps determine if there’s a statistically significant association.
Survival Analysis: Kaplan-Meier curves estimate survival distributions, while the log-rank test compares relapse-free survival (RFS) between patients with high and low T cell responses.
Predictive Modeling: Receiver Operating Characteristic (ROC) analysis, using a logistic regression model, helps assess the predictive power of T cell response.
* Software Platforms: SAS v9.4 and R v4.4.3 are used for complex statistical calculations and figure generation (Figures 1 and Extended Data Figures 2-4). GraphPad Prism v9.4 complements these tools for creating visually informative graphs (Figures 1 & 2 and Extended Data Figures 5 & 6).
Commitment to Transparency and Research integrity
We are committed to upholding the highest standards of research integrity. A detailed Reporting Summary, providing further information on our research design, is available as Supplementary material 2. This transparency ensures that our methods are open to scrutiny and contribute to the advancement of cancer immunotherapy.
this detailed approach – from precise mutation identification to rigorous statistical analysis – allows us to develop personalized immunotherapy strategies tailored to your unique cancer

![Latest Guidelines & Updates – [Industry/Topic] News Latest Guidelines & Updates – [Industry/Topic] News](https://i0.wp.com/developer.apple.com/news/images/og/apple-developer-og.png?resize=150%2C150&ssl=1)







