Anna Morena D’Alise
2026-01-16 00:00:00
Trial participants
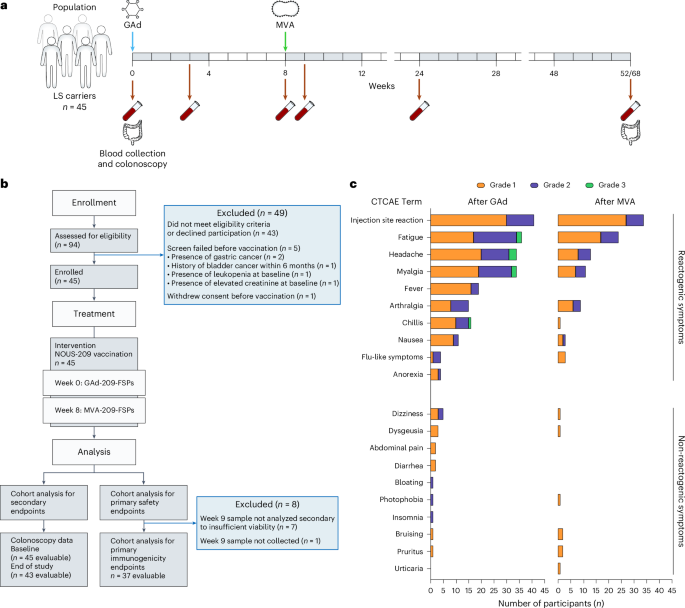
Our study population comprised individuals aged 18 years or older with a diagnosis of LS, as determined by documented carrier status of a deleterious or pathogenic or suspected to be deleterious or pathogenic (known or predicted to be detrimental or result in loss of function, respectively) germline mutation in MLH1, MSH2/EPCAM, MSH6 or PMS2, identified by a Clinical Laboratory Improvement Amendments-approved laboratory test. Consistent with the primary objectives to evaluate the safety, tolerability and immunogenicity of Nous-209 amongst healthy LS carriers, eligible trial participants had no evidence of active or recurrent invasive cancers for at least 6 months before screening and received no cancer-directed treatment (surgery, systemic therapy, hormonal therapy or radiation) within 6 months before screening. We excluded participants who had histologic evidence of high-grade dysplasia and/or invasive cancer at baseline screening. Eligible participants had adequate organ function and Eastern Cooperative Oncology Group (ECOG) performance status 0–1. Except for cardiopreventive aspirin (10 mg daily of prednisone equivalents) or other immunosuppressive medications within 14 days of study treatment. Females who were pregnant or breastfeeding or planning to become pregnant or men attempting or planning to conceive children within 6 months of the end of study treatment were excluded. Detailed inclusion and exclusion criteria are available in the study protocol (Supplementary Information).
Trial design, treatment and oversight
The trial was a phase 1b/2 single-arm, open-label, multicenter, prospective study originally designed with the coprimary endpoints of safety and immunogenicity following initial vaccination with Nous-209 monotherapy. To achieve a goal of at least 36 individuals evaluable for the primary immunogenicity endpoint, up to 45 participants were enrolled between November 2022 and November 2023 at four institutions (The University of Texas MD Anderson Cancer Center (MDACC), The University of Puerto Rico, Fox Chase Cancer Center and City of Hope) within the National Cancer Institute (NCI) iCAN PREVENT clinical trial consortium. At baseline, all participants underwent standard-of-care screening lower endoscopy (flexible sigmoidoscopy or colonoscopy). Confirmed eligible participants received initial Nous-209 vaccination as a single 1-ml IM injection of GAd20-209-FSPs (nominal concentration of 2 × 1011 viral particles per ml) at week 0 (prime), followed by a single 1-ml IM injection of MVA-209-FSPs (nominal concentration of 2 × 108 infectious units per ml) at week 8 (boost).
Following key preactivation amendments, protocol version 5 was approved for study initiation in August 2022. In October 2023, protocol version 5.3 was approved, allowing for the addition of a revaccination cohort (cohort 2) in which a subset of eligible participants who completed initial Nous-209 vaccination at week 0 and wk 8 (cohort 1) were then randomized to receive either an MVA-209-FSP IM injection at week 52 or a GAd20-209-FSP IM injection at week 52 followed by an MVA-209-FSP IM injection at week 60. Safety and immunogenicity outcomes related to cohort 2 will be reported in a future manuscript. Separate reporting of cohorts 1 and 2 was permitted by protocol.
Our study was designed and developed by academic authors in collaboration with the Division of Cancer Prevention of the NCI and Nouscom. All authors confirm that the study and analyses were conducted in accordance with the general principles of the Declaration of Helsinki and Good Clinical Practice guidelines of the International Council for Harmonization. Written informed consent was obtained from all study participants. The NCI Central Institutional Review Board and The University of Texas MDACC Institutional Review Board approved this study (protocol nos. MDA21-06-01 and 2022-0065, respectively). Safety monitoring was performed regularly by the Data Safety Monitoring Board of MDACC. The trial was registered on ClinicalTrials.gov (NCT05078866).
Study endpoints and assessments
The coprimary endpoints of the trial were safety (rate of grade 2 and 3 AEs) and immunogenicity following initial Nous-209 vaccination. Our primary safety and tolerability endpoint was assessed during the prime and boost vaccination phase (week 0 through week 9 + 7 days). AEs were monitored throughout the study (up to 52 weeks + 14 days following initial vaccination) in all participants who received at least the GAd-209-FSP vaccination at week 0 and were graded according to version 5.0 of the NCI Common Toxicity Criteria for AEs. After each vaccine injection, participants were asked to record symptom reactivity events daily on a memory aid (vaccine report card) for up to 7 days (or up to 8 days after symptom resolution). Protocol-defined injection-site reactions included pain, tenderness, erythema or redness, induration or swelling, itching and bruising, while systemic reactogenicity symptoms included fever, chills, malaise, fatigue, myalgia or muscle aches, headache, nausea, vomiting, anorexia and arthralgia or joint pain.
Our coprimary immunogenicity endpoint was assessed at week 9 and was defined as reactivity to at least one of the 16 FSP pools using an ELISpot assay. Notably, in the case of detection of reactivity pools at baseline, an increase of at least 80% in the preexisting reactivity (measured at baseline) was considered as a positive response to the vaccine.
Per protocol, evaluable participants underwent screening lower endoscopy (colonoscopy or flexible sigmoidoscopy) at week 52 ± 14 days (cohort 1) or week 68 ± 14 days (cohort 2) for standard-of-care endoscopic assessment in accordance with local institutional practices for high-risk screening populations and for collection of research biopsies. For all participants, clinical endoscopic biopsies of abnormal mucosa and/or resected polyp specimens (if any) were submitted for routine clinical pathology assessment at each participating study site. As prespecified secondary endpoints, we recorded polyp burden (count, size, histology and presence or absence of high-grade dysplasia) and neoplasia incidence. Additional secondary endpoints are detailed in the study protocol (Supplementary Information).
Participant PBMC isolation
PBMCs from whole blood were isolated at different time points and cryopreserved at each of the clinical sites before shipment to the central laboratory for immunogenicity assessment at Nouscom. To maintain the functionality of PBMCs, isolation and freezing procedures were completed within a maximum of 8 h from blood collection. PBMCs were isolated using Leucosep Bio-One polypropylene tubes (prefilled; Greiner, Merck) following the manufacturer’s instructions. Cryopreserved cells were thawed, washed, counted and rested overnight before use in immunological assays.
Peptides
A set of 976 recombinant, lyophilized peptides, with the majority of them being 15 aa in length, overlapping by 11 aa and spanning the entire sequence of Nous-209, were produced by JPT Peptide Technologies. Individual FSPs were covered by its specific pool of overlapping peptides and then arranged in 16 peptide pools for immunogenicity assessment, as described below. Lyophilized peptides were reconstituted at 40 mg ml−1 in sterile DMSO (Sigma, D2650), aliquoted and stored at −80 °C. To prepare pools 1–16, the peptides were mixed to a final concentration of 0.4 mg ml−1 for each peptide.
IFNγ ELISpot assay
IFNγ ELISpot assays were performed ex vivo in triplicate with 2 × 105 PBMCs per well in R10. PBMCs were resuspended in R10 medium, stimulated with a set of peptides designed to cover the 209 FSPs encoded by the vaccine and arranged into 16 peptide pools (P1–P16) at a final concentration of 3 µg ml−1. Cells were plated in ELISpot plates (human IFNγ ELISpot PLUS kit, Mabtech) and incubated for 18–20 h at 37 °C in a humidified CO2 incubator. At the end of incubation, the ELISpot assay was developed according to the manufacturer’s instructions. Spontaneous cytokine production (background) was measured by incubating PBMCs with medium alone, supplemented with the peptide diluent DMSO (negative control, Sigma-Aldrich), whereas CEFX (JPT Peptide Technologies), a pool of known peptide epitopes for a range of HLA subtypes and different infectious agents, was used as positive control. Results are expressed as SFCs per 106 PBMCs in stimulated cultures after subtracting the DMSO background. A response was considered positive if (1) the number of SFCs per 106 PBMCs ≥ 50 and (2) it was at least twice the DMSO background value. A subject was classified as a responder if reactivity to at least one of the 16 FSP peptide pools is induced after vaccination. If a subject exhibited preexisting reactivity to a peptide pool at baseline, the vaccine was expected to enhance this response by at least 80% in at least one of the 16 FSP peptide pools; in this case, such participants were considered as responders. NeoAg vaccine peptide pools were deconvoluted to identify immunogenic peptides by IFNγ ELISpot assays ex vivo or after in vitro stimulation. ELISpot plates were analyzed on the CTL ImmunoSpot S6 universal analyzer.
Characterization of CD4⁺ and CD8⁺ T cell responses
To characterize neoAg-induced CD4⁺ and CD8⁺ T cell responses, CD8+ T cells were selectively depleted from the total using anti-CD8 microbeads (Miltenyi Biotech, 130-045-201) following the manufacturer’s instructions. The CD8− cell population was then stimulated with either individual peptides or peptide pools (final concentration 3 μg ml−1) and T cell responses against specific peptides were assessed using an IFNγ ELISpot assay. The depletion efficiency of CD8+ T cells was confirmed by flow cytometry. T cell responses were classified as CD8⁺ mediated if a significant reduction in IFNγ spot count was observed following CD8⁺ T cell depletion. Conversely, responses were classified as CD4⁺ mediated if no substantial change in spot count was detected after depletion
Assessment of T cell immunophenotype and cytokine production by flow cytometry
PBMCs were thawed and rested for 1 h at 37 °C in R10. The samples were then incubated with anti-human Fc block (Pharmingen BD) at a 1:50 dilution in fluorescence-activated cell sorting (FACS) buffer (1× PBS and 0.5% FBS) for 20 min at 4 °C. After washing with FACS buffer, the samples were stained with HLA class I dextramer HLA-B*0801/IAKKRIKL 8-mer peptide (SPEF2 FSP) conjugated to PE at room temperature for 30 min, protected from light. The live/dead near-IR dead cell stain kit (Thermo Fisher Scientific, L10119) was added at 1:100 dilution and cells were incubated for 15 min at room temperature. The following surface staining antibodies were added: CD4 (BioLegend, clone A161A1, 357406), CD8 (BioLegend, clone SK1, 344710), CD45RA (BioLegend, clone HI100, 304142) and CCR7 (CD197; BioLegend, clone G043H7, 353226). After a 30-min incubation at room temperature, the samples were washed and resuspended in FACS buffer until acquisition. For intracellular staining, PBMCs were stimulated with the vaccine single peptide (from SPEF2, 11-mer) (4 µg ml−1), DMSO (control) and PMA/ionomycin cell stimulation cocktail (positive control; Affymetrix) in the presence of anti-human CD107a (LAMP1; BioLegend, clone H4A3, 328618). After overnight coculture, BD GolgiPlug transport inhibitors (BD, 51-2301KZ) were added. Following 3 h of incubation at 37 °C, the samples were washed with FACS buffer and incubated with Fc block for 20 min at 4 °C. The samples were then washed and stained with live/dead dye in staining buffer for 15 minutes at room temperature. For surface staining, cells were labeled with CD4 (BioLegend, clone A161A1, 357406) and CD8 (BioLegend, clone SK1, 344710) antibodies. After a 15-min incubation at room temperature, the samples were washed twice before fixation and permeabilization with CytoFix/CytoPerm (BD Cytofix/Cytoperm kit) for 15 min at 4 °C. The samples were washed twice and resuspended in 1× perm/wash buffer (BD Cytofix/Cytoperm kit) for intracellular staining with IFNγ (BioLegend, clone 4S.B3, 502532). After 30 min of incubation at room temperature, the samples were washed and then resuspended in perm/wash buffer until acquisition. Data were acquired on a BD FACS Canto II and analyzed using FlowJo (version 10.1). The gating strategies are provided in Supplementary Fig. 2.
Genetic modification of HCT116 colon cancer cell line for HLA allele and neoAg expression
To investigate neoAg-specific immune responses, we genetically modified the MMRd HCT116 colon cancer cell line, which harbors an MLH1 mutation, to express multiple HLA alleles and neoAg minigenes (MGs) derived from the CDC7 gene mutation. These MGs encode neoAg peptides of varying amino acid lengths (specifically 50-mer, 15-mer and 9-mer) designed to be presented by both MHC class I and class II alleles, as informed by previous studies7. The codon-optimized CDC7 50-mer construct was cloned into a lentiviral backbone obtained from VectorBuilder (plasmid VB240607-1533chz). The design of the construct included an N-terminal signal peptide (MSPMRVTAPRTLILLLSGALALTETWAGS), the mutated CDC7 epitope containing the HLA-A*03:01-restricted 15-mer (TSRILNLQVLKKILR) with its minimal 9-mer core (TSRILNLQV), an MHC I trafficking domain (MCLRLRTKLEKALSALFIWPQHSYKIVGIVAGLAVLAVVVIGAVVATVMCRRKSSGG) and a flexible C-terminal linker (KGGSYSQAASSDSAQGSDVSLTA). This configuration ensured efficient routing of the CDC7 construct through the secretory pathway and enhanced antigen processing and presentation for recognition by neoAg-specific T cells. HCT116 cells were transfected to express HLA-A11:02, A24:01, A03:01 and B07:02, in addition to their endogenous alleles (HLA-A02:01, A01:01, and B45:01). The transfection process followed a two-step protocol. In the first step, HLA allele expression cassettes were cloned into PiggyBac transposon vectors under constitutive promoters and cotransfected with a transposase helper plasmid into HCT116 cells using Lipofectamine 3000 according to the manufacturer’s instructions. The fluorescent reporters (red fluorescent protein, blue fluorescent protein and enhanced green fluorescent protein (EGFP)) were included to enable tracking of transgene expression and selection of stable clones by drug selection, followed by flow cytometry sorting to ensure the expression of all transgenes within single cells. In the second step, HCT116 cells carrying the HLA constructs were transduced with ready-to-use lentiviral particles generated by VectorBuilder carrying the CDC7 50-mer construct. Cells were plated 1 day before infection, exposed to viral supernatant supplemented with 8 μg ml−1 polybrene and spinoculated at 800g for 90 min at 32 °C to enhance transduction efficiency. After overnight incubation, the medium was replaced with fresh complete DMEM and cells were expanded for 5–7 days. Puromycin selection was applied to enrich for stable integrants, which were further purified by FACS on the basis of EGFP expression. Stable clones were subsequently validated by PCR and sequencing for integration, by western blot and flow cytometry for expression and by functional assays to confirm HLA surface expression and CDC7 MG presentation.
In vitro stimulation and functional validation of CDC7 neoAg contained in Nous-209
For the functional evaluation of neoAg-specific immune responses, the 15-mer peptide CDC7 (TSRILNLQVLKKILR), which is restricted to MHC I, was used for in vitro validation. The peptide was synthesized by JPT Peptide Technologies and used to stimulate PBMCs derived from LS trial participant 12 collected at baseline and at week 8 after Nous-209. PBMCs were cultured in R10 medium (RPMI 1640 with L-glutamine (Corning, 10040CV), 10% heat-inactivated FBS (HyClone, SH30070.03), 10 mM HEPES buffer (Corning, 25060-CI) and 1× penicillin–streptomycin (Corning, 30002CI)), supplemented with 330 U per ml recombinant human IL-7. Stimulation was carried out using 4 µg ml−1 CDC7 peptide per well, with concanavalin A and DMSO serving as positive and negative controls, respectively. On days 3, 7 and 10, the cells were replenished with R10 medium containing 10 U per ml interleukin 2 (IL-2). After 12 days of culture, the expanded T cells were divided into two populations; one half was used for microfluidic coculture assays to assess tumor cell-targeting potency, while the other half was subjected to IL-2 withdrawal for subsequent IFNγ ELISpot analysis. On day 13, the second population of stimulated PBMCs was seeded in triplicate (3 × 105 cells per well) into 96-well ELISpot plates precoated with human IFNγ capture antibodies. The cells were restimulated with 3 µg ml−1 CDC7 peptide and incubated for 16–20 h. IFNγ secretion was measured using the ELISpot assay, following the manufacturer’s protocol (Mabtech). SFCs were quantified using the ImmunoSpot S6 UNIVERSAL analyzer (Cellular Technology Limited).
Tumor elimination by neoAg-stimulated T cells using a microfluidic coculture system
To evaluate the tumor-targeting potency of neoAg-stimulated T cells, experiments were performed using the OrganoPlate three-lane 64 microfluidic system (MIMETAS). A collagen I extracellular matrix (ECM) gel was prepared by mixing collagen I (5 mg ml−1; AMSbio), 1 M HEPES (Thermo Fisher Scientific) and NaHCO3 (Sigma) in a 1:1:8 ratio on ice. Then, 2 μl of the gel mixture was loaded into each gel inlet and polymerized in a humidified incubator at 37 °C for 15 min. Genetically modified HCT116 cells expressing CDC7 MG derived from the CDC7 gene mutation, encoding peptides of varying amino acid lengths, were used in these experiments. Cells were labeled with the fluorescent dye CellTracker red CMTPX (Thermo Fisher Scientific) and seeded into the top perfusion inlets adjacent to the ECM gel channels at a density of 10,000 cells per µl (2 µl per chip). The plate was incubated on its side for 3–4 h to ensure cell attachment to the ECM gel. NeoAg-stimulated PBMCs were labeled with the fluorescent dye CellTracker green CMFDA (Thermo Fisher Scientific) and seeded into the bottom perfusion channels at a 5:1 E:T ratio. Following cell seeding, 50 µl of culture medium was added to all perfusion inlets and outlets to ensure complete filling of the channels without air bubbles. The coculture system was maintained on a rocker platform (14° inclination, 8-min intervals) for 48 h to enable continuous medium perfusion. After the 48-h incubation, genetically modified HCT116 cells and control cells were collected from the microfluidic chamber outlets. Migrated T cells were depleted from the collected samples using CD3 MicroBeads (Miltenyi Biotec) and column-based magnetic separation according to the manufacturer’s instructions. This process ensured the efficient removal of CD3+ T cells, leaving a purified tumor cell population for downstream analyses. Tumor cell viability was subsequently assessed using the CellTiter-Glo luminescent cell viability assay (Promega). Apoptotic cell death was quantified using a caspase 3/7 luminescence assay, performed in accordance with the manufacturer’s protocols. For viability, equal volumes of reagent and culture medium were added to samples, inducing cell lysis and generating a stable luminescent signal proportional to intracellular adenosine triphosphate content. Luminescence was recorded using a plate luminometer, expressed as relative luminescence units, normalized to untreated control wells and reported as percentage viability relative to control. For apoptosis, equal volumes of Caspase-Glo 3/7 reagent and sample were combined, resulting in caspase-mediated cleavage of a luminogenic DEVD substrate and light emission proportional to caspase 3/7 activity. Luminescence was recorded on a plate luminometer, normalized to control wells and expressed as percentage apoptosis relative to baseline.
Genomic DNA extraction and whole-exome sequencing (WES)
Genomic DNA was extracted from five serial slides from formalin-fixed paraffin-embedded (FFPE) blocks of endoscopic resections of colorectal adenomas from on-study colonoscopies. First, we deparaffinized the tissue sections with xylene and 100% alcohol. Then, tissues were collected and incubated with lysis buffer in the presence of proteinase K using the Roche microRNA isolation kit. Lysates were centrifuged for 30 min at 4 °C at 15,000 rpm and cell pellets were used for extraction of genomic DNA using the AllPrep DNA/RNA FFPE kit (Qiagen), following the manufacturer’s protocol. DNA quality was assessed using TapeStation analyzer; then, Twist exome capture, library preparation and raw sequencing were performed by the Advanced Technology Genomics Core at The University of Texas MDACC using the Illumina NovaSeqX platform. Alignment of WES data was performed using BWA-mem (version 0.7.19) with default parameters to human genome reference hg38. Duplicate reads were marked with GATK (version 4.6.2.0). Base quality recalibration was performed with GATK Apply BQSR.
Existing WES data processing
Tumor exome data were processed starting from the raw data (FASTQ files), which were downloaded from the National Center for Biotechnology Information under BioProject PRJNA954699. A preliminary quality control of the raw sequence data was performed by filtering out reads of low quality with Trimmomatic (version 0.33)33. The remaining reads were aligned on the GRCh37 human genome BWA-mem (version 0.7.17-r1188)34. Multimapping reads were filtered out using SAMtools (version 1.9)35. Optical duplicates were marked using Picard’s MarkDuplicates tool with Picard tools. DNA alignments were further optimized at regions around indels and base scores were recalibrated after the optimization step using GATK software (version 3.7)36. Frameshift mutations within the Nous-209 neoAgs were identified from aligned sequencing data (BAM files) using a lookup-based approach. A mutation was considered present if a minimum of three reads supported the variant and the variant allele frequency (VAF) exceeded 10%, consistent with previously established criteria.27
MSI status assessment
MSI status of previously published datasets7 and colorectal adenomas from on-study colonoscopies was determined using MSIsensor2 (version 0.1; https://github.com/niu-lab/msisensor2.git)37 in ‘tumor-only’ mode. The pipeline involved indexing the reference genome (hg19), scanning 2,793 MS sites and calculating MSI scores on the basis of the proportion of unstable loci. A sample was classified as MSI-H if the MSI score exceeded the predefined threshold of 20%, MSI-L if the MSI score was between 10% and 20% and MSS if the score was
MHC class I prediction
MHC class I binding affinity predictions were performed using the Immune Epitope Database and Analysis Resource MHC I prediction tool (https://www.iedb.org/). Peptide sequences of interest were analyzed for their potential to bind HLA class I molecules using the Consensus method (version 2.18) with peptide lengths of 8, 9 and 10 aa. The analysis included the prediction of binding affinities (half-maximal inhibitory concentration values) for HLA alleles. Downstream immunogenicity analyses were performed, prioritizing peptides on the basis of the results of MHC class I binding affinity predictions.
Statistical analyses
The trial was conducted using Simon’s minimax two-stage design, wherein the immunogenicity response rate was defined by the number of evaluable participants with immunogenicity by ELISpot assay among all treated participants. On the basis of prior evidence38, our primary efficacy endpoint was evaluated with respect to a predefined target immunogenicity response rate of ≥75%; by contrast, we considered a immunogenicity response rate of ≤55% to be unacceptable. In the first stage, 24 participants were enrolled and accrual halted to fully evaluate immunogenicity at week 9; with 16 or more responses observed in the first stage, additional participants enrolled in the study to reach a total of 36 evaluable participants. Upon study completion, Nous-209 vaccination was considered effective if >24 participants demonstrated immunogenicity. Under these operating characteristics, if the true immunogenicity response rate is 0.55, the probability of stopping the trial early was 83% at an expected sample size 26. Immunogenicity rates are reported with 95% exact confidence intervals evaluated using the Clopper–Pearson method. Our study applied a Bayesian toxicity monitoring plan in which treatment would be considered unsafe if the estimated rate of unacceptable toxicity (grade 3 of higher treatment-related AEs except for vaccine reactogenicity symptoms) was ≥30% with a probability of ≥70%. Assuming a β prior probability of toxicity with parameters (0.3, 0.7) and considering the Simon’s two-stage design, the trial had a 99.6% chance of stopping early if the true toxicity rate was 50% when the true response rate was 55%. SAS 9.4 was used for exploratory statistical analyses of associations between clinical and demographic factors and immunogenicity. Frequencies and percentages are reported for categorical variables. Summary statistics such as number of nonmissing observations, mean, median, s.d., minimum and maximum are provided for continuous data. The chi-squared test and Fisher’s exact test were used to evaluate the association between categorical variables and vaccine responses. Wilcoxon’s rank sum test or Kruskal–Wallis test was used to compare the distributions of continuous response variables (such as the number positive pools at specific time points), across different demographic or clinical groups. Statistical analyses for cell viability, apoptosis and cytokine secretion were conducted using Prism 10 (GraphPad Software). For cell viability and apoptosis assays, data were analyzed across the following groups: genetically modified HCT116 cells cocultured with CDC7-stimulated PBMCs (patient 12 at baseline and after Nous-209), nongenetically modified HCT116 cells cocultured with CDC7-stimulated patient 12 PBMCs at baseline and after Nous-209 (control group) and genetically modified HCT116 cells cultured alone (CDC7-HCT116 control). Statistical comparisons were performed using a two-sided unpaired t-test, with significance indicated in the figure (P values t-tests were used to compare PBMCs at baseline and after Nous-209, with comparisons to the negative control (DMSO) and results expressed as the mean ± s.d.; statistical significance was defined as P P P P P
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.