Unlocking Parkinson’s: New Research Reveals How Toxic Protein Structures Damage Brain Cells in Real-time
Parkinson’s disease, a progressive neurological disorder affecting millions worldwide, has long presented a frustrating enigma to researchers. While characterized by hallmark motor symptoms like tremors and rigidity, the underlying cause of the disease – and how brain cells succumb to its devastating effects – has remained elusive. Now, groundbreaking research from Aarhus University is offering a critical new piece of the puzzle, revealing a dynamic process of cellular attack driven by misfolded proteins. This study, published in the prestigious journal ACS Nano, doesn’t just observe the damage; it captures it happening in real-time, opening doors to earlier diagnosis and potentially more effective therapies.
The Long-Held mystery of Parkinson’s and the Role of α-Synuclein
For years, the focus in Parkinson’s research centered on large, visible protein clumps called fibrils found in the brains of affected individuals. These fibrils, formed from the protein α-synuclein, were thought to be central to the disease process. α-Synuclein is a naturally occurring protein vital for healthy brain function, playing a key role in interaction between nerve cells. Though, in parkinson’s, this protein undergoes a risky transformation, misfolding and aggregating into toxic structures.
recent evidence, and now powerfully confirmed by the Aarhus University team, points to a more insidious culprit: smaller, less visible structures called α-synuclein oligomers. These oligomers are proving to be substantially more toxic than their larger fibril counterparts, and this new research details how they inflict damage at the cellular level.
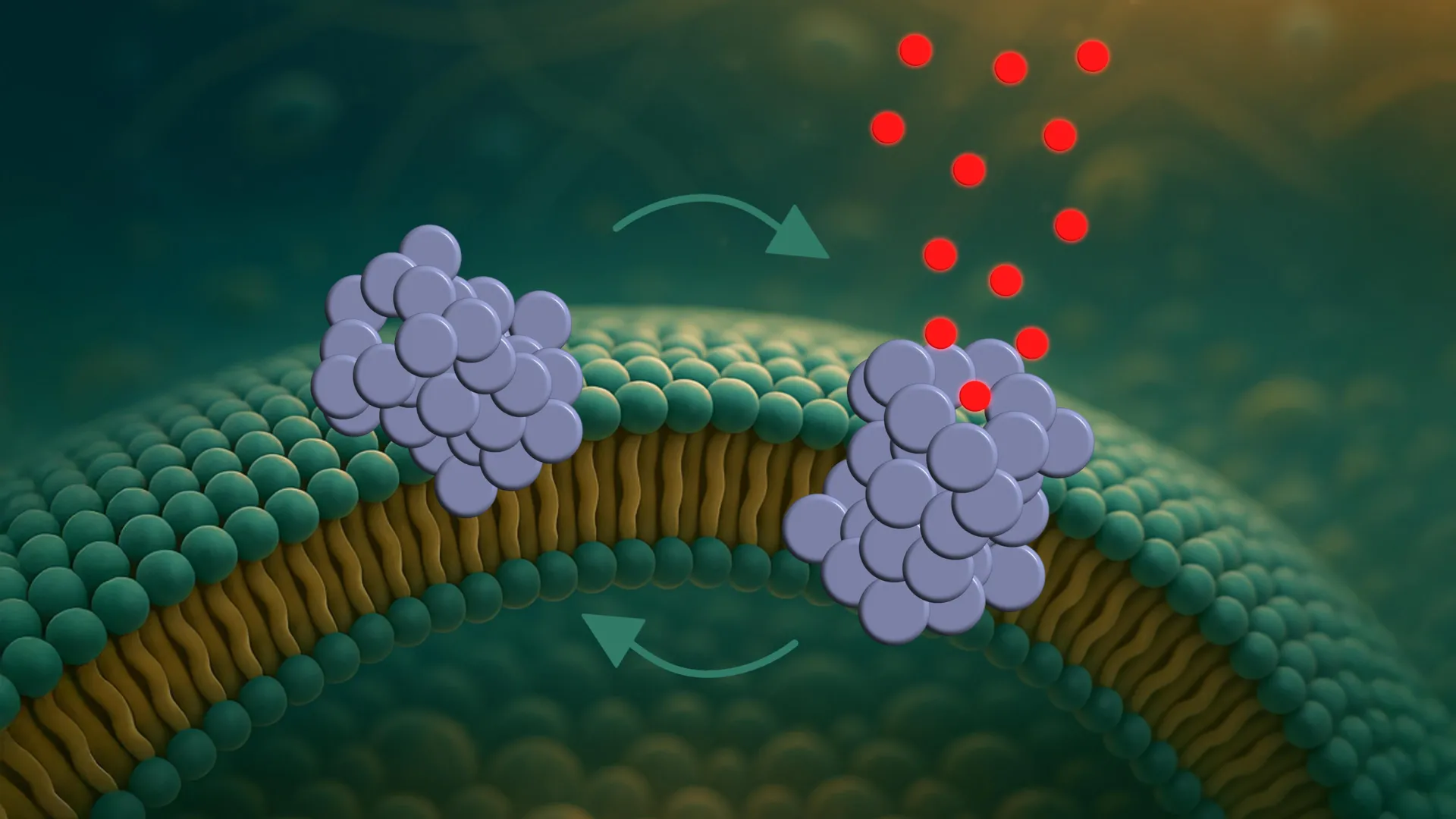
A Molecular Movie: Observing Cellular Damage in Real-Time
The breakthrough lies in a novel single-vesicle analysis platform developed by the Aarhus team. This innovative technology allows researchers to observe the interactions between individual proteins and individual vesicles - essentially, artificial bubbles mimicking cell membranes. This isn’t just a snapshot; it’s a “molecular movie in slow motion,” as described by postdoctoral researcher Mette Galsgaard Malle, who collaborated on the study with Harvard University.
What the researchers observed is a three-stage process:
- Attachment: Oligomers initially bind to the cell membrane,particularly in areas of curvature.
- Insertion: They then partially embed themselves within the membrane structure.
- Pore Formation: they assemble into dynamic pores - microscopic holes – that disrupt the cell’s delicate internal balance.
Crucially, these pores aren’t static. they constantly open and close, behaving like “tiny revolving doors,” explains PhD student and first author bo Volf Brøchner. this dynamic behavior is significant. If the pores remained permanently open, the cell would likely collapse rapidly. The intermittent opening and closing allows the cell’s internal “pumps” a window of chance to attempt to restore equilibrium, delaying – but not preventing – eventual cell death.
Implications for Diagnosis and Treatment: Nanobodies and Mitochondrial Targeting
This real-time observation of pore dynamics is a game-changer. It provides a concrete target for therapeutic intervention and offers a new avenue for early disease detection. The team has already begun exploring the potential of nanobodies - small antibody fragments - designed to bind to these oligomers. While initial tests showed the nanobodies didn’t directly block pore formation, they demonstrated promise as highly selective diagnostic tools, potentially allowing for the detection of oligomers at the very earliest stages of the disease – long before significant neuronal damage occurs. Currently, parkinson’s is often diagnosed only after ample brain cell loss has already taken place.
Further investigation revealed another critical detail: these pores don’t form randomly. They preferentially emerge in membrane types resembling those found in mitochondria, the cell’s powerhouses. This suggests that mitochondrial dysfunction may be an early event in the Parkinson’s disease process, and that damage may originate within these vital cellular components.
The Road Ahead: From model systems to Biological Realities
While these findings are incredibly promising, the researchers emphasize the study was conducted using simplified model systems. The next crucial step is to replicate these results in living cells and, ultimately, in biological tissue, where a far more complex interplay of factors is at play.
“We created a clean experimental setup where we can measure one thing at a time. That’s the strength of this platform,” explains malle.”But now we need to take the next step and investigate what happens in more complex biological systems.”
This research represents a significant leap forward in our understanding of Parkinson’s disease. By visualizing the molecular mechanisms of cellular damage in real-time, the Aarhus University team has not only illuminated a critical pathway in disease progression but also provided a powerful