Renee R. Fedak
2026-01-09 00:00:00
Preclinical in vitro experiments
Generation of CAR cells and evaluation of CAR expression
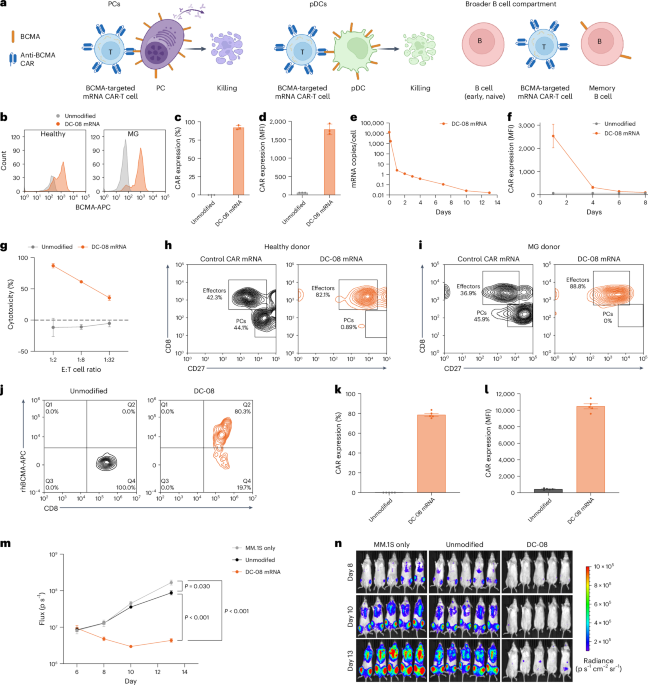
Primary CD8+ T cells were isolated from PBMCs of HDs or patients with MG using the Miltenyi Biotec CD8 MicroBeads kit, following the manufacturer’s positive selection protocol. In brief, single-suspension PBMCs were incubated with CD8 MicroBeads and passed through an LS column placed in a magnetic field; unlabeled cells were washed away, and the positively selected CD8+ T cells were eluted. Selected T cells were cultured in X-VIVO 15 medium (Lonza) supplemented with 5% human AB serum (GeminiBio) and GlutaMAX (Thermo Fisher Scientific) at a density of 1 × 106 cells per milliliter. Recombinant human IL-7 and IL-15 (Proteintech) were added at 100 ng ml−1 each, and 25 ng ml−1 OKT3 (Takara Bio, anti-CD3 monoclonal antibody) was added on culture day 0 to initiate T cell activation and expansion. Culture was monitored every 3–4 days for cell density and viability. Cytokine-supplemented media were added as needed to maintain cell culture density at 1 × 106 cells per milliliter. After culture, activated T cells were transfected with Descartes-08 mRNA. After transfection, CAR expression was assessed by flow cytometry using APC-conjugated recombinant human BCMA (BCMA-APC; ACROBiosystems). Transfected cells were washed with FACS buffer (PBS + 2% FBS), stained with propidium iodide and BCMA-APC for 20 minutes at room temperature in the dark, washed again and analyzed by flow cytometry (Cytek Biosciences, Guava easyCyte 12HT). Data analysis was performed using guavaSoft version 3.3 and FlowJo version 10.8.1 software.
In vitro cytotoxicity assay
Target cells
For in vitro cytotoxicity assays, target cells included MM.1S-GFP cells, generated by lentiviral transduction of a GFP cassette into MM.1S cells (American Type Culture Collection) and autologous plasma cells differentiated from memory B cells in PBMCs obtained from HDs or individuals with MG. For in vitro experiments, sample sizes were chosen to achieve statistical power based on historical study data5,60.
Memory B cell isolation and plasma cell differentiation
Memory B cells were isolated from frozen PBMCs using the Memory B Cell Isolation Kit (Miltenyi Biotec) via magnetic-activated cell sorting (MACS) according to the manufacturer’s protocol. Enrichment purity (>90%) was confirmed via flow cytometry using CD19 (BioLegend, clone 4G7) and CD27 (BioLegend, clone O323). Enriched B cells were CD19+ and CD27+. Plasma cells were differentiated from memory B cells according to the method by Jourdan et al.61,62. Enriched memory B cells were expanded in complete IMDM (Gibco) supplemented with FBS (VWR), GlutaMAX (Life Technologies), CpG 2006 (Invivogen), soluble CD40L (R&D Systems), HA-tag monoclonal antibody (R&D Systems) and IL-2, IL-10 and IL-15 (PeproTech) at a culture density of 0.3 × 106 cells per milliliter in 24-well plates for 4 days. Expanded B cells were assessed for proliferation and phenotype and then cultured in plasmablast differentiation media (supplied with IL-2, IL-10 and IL-15 (PeproTech) and IL-6 (Miltenyi Biotec)) for 3–4 days at 0.5 × 106 cells per milliliter. Plasmablasts were then differentiated to plasma cells using differentiation media (IL-6, IL-15 and IFNα, all Miltenyi Biotec) for an additional 3–4 days. Final plasma cells were phenotyped via flow cytometry using CD19 (BioLegend, clone 4G7), CD27 (BioLegend, clone O323), CD38 (BioLegend, clone HIT2), CD138 (Thermo Fisher Scientific, clone 300506) and CD269 (Miltenyi Biotec, clone REA315). Example gating strategies are shown in Supplementary Fig. 1.
Cytotoxicity assay
MM.1S-GFP cells were co-cultured with Descartes-08 or unmodified CD8+ T cells at an effector:target (E:T) ratio of 1:2, 1:8 and 1:32 in a 96-well V-bottom plate. After overnight incubation, plates were centrifuged (400g, 5 minutes), and cell pellets were stained with propidium iodide (Sigma-Aldrich) for viability before proceeding with flow cytometry (Cytek Biosciences, Guava easyCyte 12HT). Cytotoxicity was quantified, and percent killing was calculated as the percentage decrease in viable MM.1S-GFP cells relative to control wells (MM.1S-GFP cells alone without effector cells). Differentiated plasma cells were co-cultured with autologous Descartes-08 or control CD8 T cells at an E:T ratio of 2:1 in 200 μl of complete IMDM media in 96-well U-bottom plates. After overnight incubation, plates were centrifuged (400g, 5 minutes). Cell pellets were stained with propidium iodide for viability, CD8-FITC (Beckman Coulter, clone B9.11) for Descartes-08 identification and CD27-PE (BioLegend, clone O323) to identify plasma cells. Cytotoxicity was quantified by flow cytometry (Cytek Biosciences, Guava easyCyte 12HT), and percentages of effector cell and target cell populations were analyzed and plotted using FlowJo version 10.8.1 software. Example gating strategies are shown in Supplementary Fig. 2.
Cytokine analysis
Supernatant samples from MM.1S-GFP and Descartes-08 co-culture were used for LEGENDplex Human CD8/NK Panel (13-plex) bead array (BioLegend), conducted according to the manufacturer’s instructions. In brief, standards and supernatant samples were incubated with fluorescence-encoded beads precoated with cytokine-specific capture antibodies in a 96-well V-bottom plate. After incubation, the plate was washed, and detection antibodies were added to form a sandwich complex. Streptavidin-PE was then added to bind biotinylated detection antibodies, generating a fluorescent signal proportional to the concentration of each analyte. Fluorescent signal was analyzed using a flow cytometer (Cytek Biosciences, Guava easyCyte 12HT), and data were processed using the LEGENDplex Data Analysis Software Suite (BioLegend). Cytokine concentrations were interpolated from standard curves.
In vitro pharmacokinetics
For in vitro pharmacokinetics, thawed Descartes-08 cells were incubated at 37 °C in complete medium and sampled at indicated timepoints for assessment of CAR protein expression and Descartes-08 RNA content. CAR protein expression was evaluated as described above. Descartes-08 mRNA was evaluated by qRT–PCR. Total RNA was extracted from Descartes-08 using an miRNeasy kit (Qiagen) according to the manufacturer’s protocol. The RNA concentration and purity were assessed using a NanoDrop spectrophotometer. Reverse transcription was performed using SuperScript IV with oligo(dT) primers (Thermo Fisher Scientific) to synthesize cDNA. Quantitative PCR was performed using SYBR Green Master Mix (Thermo Fisher Scientific) on a real-time PCR system (Applied Biosystems, ABI PRISM 7000). Gene-specific primers were used to amplify the target gene, and expression levels were normalized to housekeeping gene GAPDH. Sample sizes were chosen to achieve statistical power based on historical study data60.
Preclinical in vivo experiments
Animals
Female NSG mice (NOD.Cg-PrkdcscidIl2rgtm1wJ1/SzJ), aged 6–8 weeks, were obtained from The Jackson Laboratory and housed in a specific pathogen-free facility at Noble Life Sciences. Mice were maintained in individually ventilated microisolator cages with sterilized bedding, water and food. The vivarium environment was continuously monitored to ensure temperature (18–22 °C), humidity (35–65%) and a 12-hour light/dark cycle. All animals were monitored twice daily for clinical signs of illness or distress. Animals exhibiting signs of severe morbidity, including inability to feed or drink or more than 20% body weight loss, were humanely euthanized in accordance with institutional guidelines. All procedures were approved by the Institutional Animal Care and Use Committee of Noble Life Sciences and conducted under the oversight of a licensed veterinarian and in compliance with all applicable local, state and federal guidelines. The institution holds an active Office of Laboratory Animal Welfare (OLAW) assurance. Sample sizes were chosen to achieve statistical power based on historical study data60. The study shown is one representative study of more than five analogous experiments with similar results.
Tumor engraftment
On study day 0, for tumor engraftment, NSG mice were injected intravenously via the tail vein with 2 × 106 MM.1S-Fluc cells stably expressing firefly luciferase, a gift from Y. S. Tai (Dana-Farber Cancer Institute). On study day 6, tumor burden was assessed by bioluminescence imaging (BLI) using the IVIS Spectrum Imaging System (PerkinElmer) after intraperitoneal injection of D-luciferin (150 mg kg−1). Animals were then randomized into treatment groups based on tumor signal intensity to ensure equal tumor burden across testing groups. IVIS imaging was analyzed using Living Image version 4.7.3.20616.
Test article injection and assessment
On study day 7, mice were treated via tail vein intravenous injection with either 10 × 106 unmodified CD8 T cells or Descartes-08, with a group where no test articles were injected (MM.1S-Fluc-alone group) as control. Malignant PC (multiple myeloma) progression was monitored by BLI through day 13. Body weight and clinical observations were recorded throughout the study to assess treatment tolerability and animal health.
PBMC analysis post-test article injection
On study day 8, 100 µl of peripheral blood was collected from each animal via submandibular bleed into EDTA-coated tubes to prevent coagulation. Red blood cells were lysed using ACK lysis buffer, followed by quenching via high-serum buffer (X-VIVO 15 media + 25% FBS) and then washing in FACS buffer (PBS + 2% FBS + NaAzide). Single-suspension cells were stained with propidium iodide as well as fluorochrome-conjugated antibodies specific to human CD45 (BioLegend), CD8 (BioLegend) and BCMA-APC (ACROBiosystems) for 30 minutes at 4 °C in the dark. After FACS buffer washing, samples were acquired on a flow cytometer (Agilent Technologies, NovoCyte 3000), and data were analyzed using Agilent NovoExpress version 1.6.1 software. Live cells were gated based on forward and side scatter and viability dye (propidium iodide) exclusion, followed by gating on human CD45+CD8+ events to identify CAR+ cells.
Clinical trial
MG-001 part 3 was a prospective, double-blind, multicenter, randomized placebo-controlled phase 2b trial evaluating the safety and clinical activity of Descartes-08, RNA-engineered anti-BCMA CAR-T, in adults with generalized MG. Of the 45 patients who met the inclusion criteria, 36 were eligible to be randomized after meeting manufacturing specifications, of whom 20 were assigned to the active Descartes-08 arm, and 16 were assigned to the placebo arm. All participants underwent leukapheresis and manufacturing of autologous CAR-T product. A single apheresis was sufficient to reach the target dose of 52.5 × 106 viable CAR+ cells per kilogram ± 45% for each of six once-weekly infusions. Starting on day 1, participants received the six once-weekly doses of Descartes-08 or placebo. One of the participants randomized to Descartes-08 withdrew from the study for reasons other than safety or efficacy after two infusions, and subsequent labs were not analyzed; therefore, 35 total randomized patients were analyzed in the correlative/mechanistic study. Clinical efficacy was evaluated through determination of MGC and MG-ADL scores and a clinical response threshold set at a ≥5-point drop on the MGC scale. Full details are provided in the companion paper6. Differences among the data available for the clinical efficacy (modified intention-to-treat) population with respect to clinical assessment visits versus sample availability (for example, quantity and timepoints) prevented exact alignment between the two datasets in all cases.
The study was conducted in accordance with the principles of the Declaration of Helsinki, Good Clinical Practice guidelines and applicable governmental regulatory standards. Independent institutional review boards provided written approval of the protocol and amendments. All participants provided written informed consent. The protocol is provided in the companion paper6. Sex was self-reported by participants. There were no sex-based analyses prespecified, and none was conducted post hoc due to small sample size.
Manufacturing of Descartes-08
Participants underwent leukapheresis to isolate PBMCs, which were subsequently processed and manufactured into Descartes-08 under Good Manufacturing Practice guidelines. Descartes-08 product characterization was performed according to validated release testing methods. Viability testing used an automated cell counter (Nexcelom Bioscience, K2 Cellometer) to detect viable and non-viable cells stained with acridine orange and propidium iodide, respectively. Characterization of CD8 expression and CAR expression was performed by staining of cells with CD8-APC (Beckman Coulter, clone B9.11) and recombinant human BCMA (ACROBiosytems), respectively, and analysis by flow cytometry.
Clinical biomarker analysis
Blood sample collection
Patient research samples were collected throughout Descartes-08 treatment after obtaining written informed consent. A variety of blood products and derivatives was collected, including PBMCs, serum and Tempus tube stabilized RNA. Serum was processed from peripheral whole blood using clot activator Vacutainer collection tubes. Serum was stored at −80 °C until ready for use. PBMCs were isolated from peripheral whole blood by Ficoll gradient separation. Isolated PBMCs were resuspended in a final concentration of 7% DMSO and cryopreserved until ready for use.
Flow cytometry: product analysis (CAR expression, CD8)
Flow cytometry was used to understand phenotypes of Descartes-08 products. Samples of Descartes-08 product lots were retained for exploratory use and stored at −80 °C prior to analysis. Cells were thawed and washed in a high-protein medium. Cells were stained with LIVE/DEAD Fixable Aqua Dead Cell Stain (Thermo Fisher Scientific) and washed prior to cell subset staining. Samples were then resuspended in a Fc receptor blocking solution (BioLegend) and left to incubate at room temperature for 10 minutes. Panel-specific antibodies were added to blocked product cells and incubated at 4 °C for 30 minutes. Stained cells were washed and resuspended in 200 µl of FACS buffer (DPBS + BSA + NaAzide). At least 50,000 events were recorded. All flow cytometry experiments were conducted using the Agilent NovoCyte 3000 flow cytometer. All experimental analysis was performed in Agilent NovoExpress version 1.6.1 software. Memory T cell populations were defined as follows: TCM CD3+CD4−CD8+CD28+CD45RO+, TEM CD3+CD4−CD8+CD28−CD45RO+, TSCM CD3+CD4−CD8+CD28+CD45RO− and TTE CD3+CD4−CD8+CD28−CD45RO−. Cell activation was characterized by co-expression of CD39, LAG-3 and PD-1 on CD3+ cells, as determined by fluorescence minus one (FMO) and isotype controls. Representative gating schemes were prepared using FlowJo version 10.8.1 software. Gating strategies are shown in Supplementary Figs. 4 and 5. All product phenotyping data were normalized to percent of CD3+ T cells ((count in gate X / count in CD3+ T cell gate) × 100). All data points presented are from single measurements.
Flow cytometry: PBMC phenotyping
Flow cytometry was used to understand changes in cell composition before and after Descartes-08 therapy. Cryopreserved PBMCs were thawed and washed in a high-protein medium. PBMCs were first stained with LIVE/DEAD Fixable Aqua Dead Cell Stain (Thermo Fisher Scientific) and washed prior to cell subset staining. PBMCs were then resuspended in an Fc receptor blocking solution (BioLegend) and left to incubate at room temperature for 10 minutes. Panel-specific antibodies were added to blocked PBMCs and incubated at 4 °C for 30 minutes. Stained cells were washed and resuspended in 200 µl of FACS buffer (DPBS + BSA + NaAzide). At least 40,000 events were recorded for T cell subset and TBNK panels. At least 100,000 events were recorded for B cell and dendritic cell panels due to low frequencies of populations of interest. All flow cytometry experiments were conducted using the Agilent NovoCyte 3000 flow cytometer. All experiment analysis was performed in Agilent NovoExpress version 1.6.1 software. Cell populations were defined as per the following table with gating strategies shown in Supplementary Figs. 6 and 7 (prepared using FlowJo version 10.8.1 software). The full antibody list is provided in Supplementary Table 1. Flow cytometric definition of immune cell populations is provided in Supplementary Table 2.
BCMA and CD86 surface protein expression were measured using median fluorescence intensity (MFI) on single-parameter histograms. HD ranges for circulating peripheral blood and pDC frequencies were obtained from literature10,11. Other data on HDs were obtained by flow cytometric analysis of PBMCs from a panel of HDs (demographics provided in Supplementary Table 3). All CD8+ T memory, CD4+ T memory, T helper and TBNK data were normalized to percent of live cells ((count in gate X / count in live cell gate) × 100). pDC data were normalized to both percent of live cells ((count in pDC gate / count in live cell gate) × 100) and percent of dendritic cells ((count in pDC gate / count in dendritic cell gate) × 100). Peripheral blood data were normalized to percent of B cells ((count in peripheral blood gate / count in CD19+ gate) × 100). Data were excluded for peripheral blood quantitation and BCMA MFI measurements if peripheral blood count was less than three events. For longitudinal analysis, change in cell frequency was calculated using fold change from screen (normalized population frequency at month X / normalized population frequency at screen). Change in MFI was calculated using delta change from screen (MFI at month X − MFI at screen). All data points presented are from single measurements.
RNA isolation and cDNA synthesis
Whole blood was collected at clinical sites in Tempus RNA preservation tubes (Thermo Fisher Scientific) and stored at −80 °C until processing. RNA extraction was performed using the Tempus Spin RNA Isolation Kit (Thermo Fisher Scientific) and used to generate cDNA using the SuperScript IV cDNA Synthesis Kit and oligo-dT primers (Thermo Fisher Scientific), according to the manufacturer’s instructions.
Analysis of circulating RNA from CAR-T cell product by quantitative PCR
Measurement of Descartes-08 CAR RNA in whole blood was performed by qRT–PCR using gene-specific primers on cDNA extracted from Tempus whole blood tubes. Reactions were set up using a hydrolysis probe assay (forward primer: ATGGATGGGCTGGATCAACA; reverse primer: GGTGTCCTCGTACTTCAGGT; hydrolysis probe: /{5′FAM}/ACCAGGGAGCCCGCCTACGC/{3′TAMRA}, all from Integrated DNA Technologies) and TaqMan Universal Master Mix II (Thermo Fisher Scientific). Real-time quantitative PCR was performed on an AriaMx Real-Time PCR System (Agilent Technologies). A standard curve prepared with linearized plasmid containing the amplicon was used to quantitate copy number of cDNA in each reaction, and values were normalized to circulating quantity in blood. Reactions, standard curve and no template control (NTC) were performed in triplicate and averaged for analysis. Values lower than the lower limit of quantification (LLOQ) were substituted with 2 for the purpose of data display.
PhIP-Seq
All PhIP-Seq was performed similarly to a previously published multichannel protocol:
https://www.protocols.io/view/derisi-lab-phage-immunoprecipitation-sequencing-ph-czw7x7hn?step=14.1
As previously described63, this human peptidome library consists of a custom-designed phage library of 731,724 unique T7 bacteriophages each presenting a different 49-amino-acid peptide on its surface. Collectively, these peptides tile the entire human proteome including all known isoforms (as of 2016) with 25-amino-acid overlaps. One milliliter of phage library was incubated with 1 µl of human serum (from each of the n = 279 samples) overnight at 4 °C and immunoprecipitated with 25 µl of 1:1 mixed protein A and protein G magnetic beads (Thermo Fisher Scientific, no. 10008D and no. 10009D). These beads were then washed, and the remaining phage–antibody complexes were eluted in 1 ml of Escherichia coli (EMD Millipore, BLT5403) at 0.5–0.7 optical density and amplified by growing in a 37 °C incubator. This new phage library was then reincubated with the same individual’s serum, and the previously described protocol was repeated. DNA was then extracted from the final phage library, barcoded and PCR amplified, and Illumina adaptors were added. Next-generation sequencing (NGS) was then performed using an Illumina sequencer to a read depth of approximately 1 million per sample. To control for background binding of phage directly to protein A or protein G, we also performed the identical experiment but did not include any antibody or human samples. We performed 79 of these ‘mock-IPs’.
Analysis of PhIP-Seq
As previously described, NGS reads from FASTQ files were aligned at the level of amino acids using RAPSearch2. All human peptidome analysis was performed at the gene level, in which all reads for all peptides mapping to the same gene were summed, and 0.5 reads were added to each gene to allow inclusion of genes with zero reads in mathematical analyses. Within each individual sample, reads were normalized by converting to the percentage of total reads. To normalize each sample against background non-specific binding, a fold change over mock-IP was calculated by dividing the sample read percentage for each gene by the mean read percentage of the same gene for the 79 mock-IP A/G bead-only controls. This fold change signal was then used for side-by-side comparison between samples and cohorts. As previously described14, overall changes in the autoreactome across time were determined by comparing the complete fold change over mock-IP signal across all genes in each individual against each other sample using a Pearson correlation. The Pearson correlation coefficient r values were then used for downstream quantitative analyses.
Evaluation of AChR titers, vaccine titers and total immunoglobulin titers
Autoantibody titers, immunization titers and immunoglobulin levels were measured in patient serum by a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory (Quest Diagnostics). AChR antibody titers were measured by radioimmunoassay. Diphtheria, tetanus, measles, mumps, rubella and varicella zoster virus antibody titers were measured by immunoassay. Neisseria meningitidis antibody titers were measured by a multi-analyte immunodetection assay. An immunoglobulin panel (IgA, IgG and IgM) was measured by an immunoturbidimetric assay. If the assay resulted in a value below the LLOQ, LLOQ / 2 was used for calculations of relative change from baseline for that data point. If the assay resulted in a value above the upper limit of quantification (ULOQ), ULOQ was used for calculation of relative change from baseline for that data point.
Analysis of inflammatory proteins via the Olink Target 96 Inflammation assay
Proteomic analysis was performed on patient serum using the Olink Target 96 Inflammation assay, which measures the relative abundance of 92 proteins per sample. Serum was isolated from whole blood collected from Descartes-08 patients at clinical sites and was prepared, stored and handled according to the manufacturer’s instructions. Samples were randomized on a 96-well plate prior to analysis, and 16 bridging samples (independent of the internal controls) were used per plate to combine multiple runs into a single experiment. Sample analysis was performed at Psomagen on an Olink Q100 instrument. All samples analyzed passed standard quality control metrics, and, therefore, zero samples were excluded from analysis. Data processing and preliminary analysis were performed using Olink NPX Signature version 2.0.2 software. Data from the Olink assay were quantified as NPX values (an arbitrary relative quantitation unit on a log2 scale). Patient samples were analyzed at month 1, month 3 and month 12. For two patients without month 3 samples available (one treated with Descartes-08 and one treated with placebo), data were imputed from month 2 samples. For longitudinal analysis of relative abundance of the 92 assayed proteins, baseline NPX values were subtracted from longitudinal NPX values (that is, ΔNPX = NPXtimepoint 2 – NPXtimepoint 1).
Statistical analysis of Olink Target 96 Inflammation results
To assess statistical significance, a linear mixed-effects model (lmer) was performed for each protein using the Olink Analyze package, an R package from Olink that provides tailored statistical analysis of standard Olink data outputs, to analyze how different variables interact to impact longitudinal trends. The linear mixed model used followed the general structure of NPX ~ A × B + (1 | ID), where NPX is the output of the Olink assay, A and B are fixed effects and ID accounts for random effects and is an array of unique identifiers for each patient. To analyze the effects of leukapheresis, NPX values at day 1 and screen timepoints were entered into the model, and timepoint was used as the fixed variable. To analyze the effects of treatment with Descartes-08 compared to placebo, NPX values at month 3 and screen timepoints were entered into the model, and treatment and timepoint were used as the fixed effects. To analyze the effects of treatment at timepoints without a corresponding placebo timepoint, NPX values at month 1 or month 12 and screen timepoints were entered into the model, and timepoint was used as the fixed variable. To analyze the effects of treatment within subgroups defining clinical response or prior treatment with biologics, NPX values at month 1, month 3 or month 12 and screen timepoints were entered into the model, and timepoint and clinical response or biologics treatment, respectively, were used as fixed variables.
RNA-seq analysis
Transcriptomic analysis via RNA-seq
Transcriptomic analysis at screen and month 3 timepoints was performed on mRNA from whole blood with RNA-seq. First, mRNA was extracted followed by cDNA synthesis from stabilized whole blood (see ‘RNA isolation and cDNA synthesis’). NGS libraries were prepared using the Illumina TruSeq stranded mRNA library kit per the manufacturer’s protocol followed by sequencing on an Illumina NovaSeq X Plus system at 40 million paired reads per sample. Library preparation and sequencing were performed at Psomagen.
Transcriptomic analysis via scRNA-seq
Transcriptomic analysis at the single-cell level was performed on PBMCs using scRNA-seq. In brief, cDNA and NGS libraries were prepared from cryopreserved PBMCs isolated from whole blood (see ‘Blood sample collection’) and were processed with Chromium Next GEM Single Cell 5′ HT Reagent Kits v2 per the manufacturer’s protocol. The NGS libraries were sequenced on an Illumina NovaSeq X Plus system at a minimum of 20,000 reads per cell. Sequencing was performed at Psomagen.
scRNA-seq data processing and quality control
scRNA-seq data from the Descartes-08 MG-001 clinical trial were processed using the Seurat package (version 5.0+)64 in R (version 4.3+). For each sample, raw count matrices were assessed for quality at the cellular and gene level. Cell-level metrics included the number of unique molecular identifiers (UMIs), the number of detected genes and the percentage of reads mapping to mitochondrial and ribosomal genes. Cells were filtered based on established thresholds for these metrics. To ensure data integrity, ambient RNA contamination was estimated and removed using SoupX65, and potential cellular doublets were identified and excluded using scDblFinder66. Filtered datasets from all samples were then merged for integrated downstream analysis.
Normalization, clustering and cell type annotation of scRNA-seq results
The integrated dataset was normalized and variance stabilized using the SCTransform method67. To mitigate technical batch effects between samples, we applied Harmony’s68 integration workflow. Principal component analysis was performed on the top 2,000 highly variable genes, and the first 30 principal components were used for UMAP visualization and for constructing a nearest neighbor graph.
Cell clustering was performed using the Leiden algorithm at various resolutions with a final resolution selected that yielded biologically interpretable clusters. Cells were annotated with Azimuth and the human PBMC reference dataset69 using a two-tiered system based on the expression of canonical marker genes. Level 1 annotation identified broad immune cell categories (for example, T cells, B cells and monocytes), whereas Level 2 provided more granular subtypes (for example, CD4+ naive T cells, CD14+ monocytes, pDCs and plasmablasts).
Pseudobulk data aggregation of scRNA-seq results
To facilitate differential gene expression (DGE) analysis, single-cell expression profiles were aggregated into pseudobulk samples. Raw UMI counts for each gene were summed for each cell type at both tiers within each individual sample. Sample-level metadata, including treatment condition, timepoint and patient group, were retained for each resulting pseudobulk profile. Only sample–cell type combinations exceeding a minimum cell count threshold were included in the final pseudobulk dataset.
DGE and functional enrichment analysis of RNA-seq and scRNA-seq results
DGE analysis of pseudobulk counts was conducted using DESeq2 (version 1.40+)70. The statistical model was designed to assess the effects of treatment, timepoint and patient subgroup, including interaction terms (for example, treatment × timepoint). We performed pairwise comparisons between treatment groups at specific timepoints and within defined patient subgroups. Genes were considered differentially expressed if they met a false discovery rate (FDR) threshold 2 fold change > 0.585 (1.5-fold change).
To interpret the biological significance of DGE results, GSEA was performed using the fgsea package (version 1.26+)71, testing for enrichment against the Molecular Signatures Database hallmark gene set collection. DGE and GSEA results were visualized using volcano plots, MA plots and heatmaps.
Cellular composition analysis of scRNA-seq results
Changes in cell type proportions across experimental conditions were analyzed using sccomp72, a package specifically designed for compositional data. A β-binomial regression model was employed to account for overdispersion, with a hierarchical structure to incorporate patient-level random effects. The model included fixed effects for treatment, timepoint and patient group as well as their interactions. Significance of compositional changes was determined using Bayesian inference, with results reported as effect sizes and 95% credible intervals.
Software and statistical analysis of RNA-seq and scRNA-seq results
All analyses were conducted in R (version 4.3+) using Bioconductor (version 3.17+). Key packages for data processing and visualization included ggplot2, ComplexHeatmap and patchwork. Gene set databases were accessed via msigdbr (version 7.5+), and functional analysis was supported by clusterProfiler (version 4.8+).
TCR variable β chain sequencing
Immunosequencing of the CDR3 regions of human TCRβ chains was performed using Adaptive Immunosequencing (Adaptive Biotechnologies). Complementary DNA was amplified in a bias-controlled multiplex PCR, followed by high-throughput sequencing. Sequences were collapsed and filtered in order to identify and quantitate the absolute abundance of each unique TCRβ CDR3 region for further analysis as previously described73,74,75. Data are from single measurements of individual samples.
Statistical analysis of TCRβ sequencing results
Two quantitative components of diversity were compared across samples in this study. First, Simpson clonality was calculated on productive rearrangements by:
$$sqrt{{sum }_{i=1}^{R}{p}_{i}^{2}},$$
where R is the total number of rearrangements and ({p}_{i}) is the productive frequency of rearrangement i. Values of Simpson clonality range from 0 to 1 and measure how evenly receptor sequences (rearrangements) are distributed. Clonality values approaching 0 indicate an even distribution of frequencies, whereas values approaching 1 indicate an increasingly asymmetric distribution in which one to a few clones are present at high frequencies. Second, downsampled rearrangements were calculated as the number of unique productive rearrangements in a sample after computationally downsampling to a common number of templates to control for variation in sample depth. Repertoires were randomly sampled without replacement five times, and the mean number of unique rearrangements was reported as the final value.
Clonal expansion was calculated following the same principles described previously76. To account for variation in total template count between samples, all repertoires were computationally downscaled to 100,000 templates while maintaining the relative abundance of each rearrangement. Statistical tests were performed on the downscaled template counts. Combined template counts for each rearrangement with a combined total count of at least five across the two samples being compared are treated as a fixed number of ‘trials’ in a two-sided, one-sample chi-squared test. This test gives the probability, P, of the observed template counts in each sample under the null hypothesis that these templates are evenly distributed between the two samples, relative to their respective repertoire sizes (that is, the total productive template count of each sample). Rearrangements that are more unequally distributed relative to this expected proportion will result in a lower probability. A rearrangement is considered differentially abundant if it satisfies two criteria: (1) P 77 and (2) the rearrangement has a frequency at least two-fold higher in one sample than in the other.
Statistical analysis of clinical biomarker data
In figures, box-and-whisker plots show the median, interquartile range and full range of data along with individual data points. For preclinical in vivo data, P values were calculated using log-transformed data and two-way repeated-measures ANOVA with Geisser–Greenhouse correction. For clinical biomarker analysis, unless otherwise noted, comparisons of placebo to Descartes-08 were performed using a two-sided unpaired t-test, and comparisons within each treatment group were performed using a two-sided paired t-test. Unless indicated otherwise, non-normally distributed data were log transformed before performing statistical analyses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.