Guilherme T. Ribas
2026-01-08 00:00:00
Kidney xenotransplantation and sample collection
An EGEN-2784 gene-edited pig kidney was transplanted into a 62-year-old male patient with end-stage kidney disease, recently described in ref. 5. A Yucatan miniature pig was genetically engineered with 69 genomic modifications. These included the deletion of three major glycan antigens, the inactivation of porcine endogenous retroviruses and the insertion of seven human transgenes (TNFAIP3, HMOX1, CD47, CD46, CD55, THBD and EPCR). The immunosuppressive protocol included antithymocyte globulin (ATG, 1.5 mg kg−1 on days −2 and −1), rituximab (anti-CD20, 1,000 mg on day −3), Fc-modified anti-CD154 monoclonal antibody (tegoprubart, 20 mg kg−1 on days −3, −2, −1, 1, 3, 7 and weekly thereafter) and anti-C5 antibody (ravulizumab, 3,330 mg on days −1, 7 and 28). This regimen was combined with standard maintenance immunosuppression consisting of tacrolimus, mycophenolic acid (540 mg twice daily) and glucocorticoids starting on day 0. On post-transplant day 8, a biopsy revealed Banff grade 2A TCMR without signs of thrombotic microangiopathy or antibody-mediated rejection. Treatment included a 500-mg pulse of methylprednisolone and anti-IL-6 receptor monoclonal antibody (tocilizumab, 8 mg kg−1). Additional 500-mg methylprednisolone pulses were given on days 9 and 10, along with ATG (1.5 mg kg−1). Tacrolimus and mycophenolic acid doses were increased. Due to C3 deposition seen in the biopsy, pegcetacoplan, a targeted C3 and C3b inhibitor, was administered. However, as there was no evidence of antibody-mediated rejection, no further doses of tocilizumab were given.
Peripheral blood was collected for serum and plasma at pretransplantation (day −3), post-xenotransplantation (days 13, 20, 26, 33 and 51) and during suspected rejection (day 7 post-transplantation). Blood was collected in heparinized tubes for PBMCs isolation at pretransplantation (day −3) and post-xenotransplantation (days 13, 20, 26 and 33). Blood was also collected in Paxgene tubes for RNA profiling (days −3 and 7) and post-xenotransplantation (day 26). The sample collection schedule is shown in Fig. 1a. The patient provided written informed consent, as approved by the MGH Human Research Committee (IRB no: 2023P003631).
Recipient PBMC and serum isolation
PBMCs were isolated using density gradient centrifugation using Ficoll-Paque solution (GE HealthCare), counted and cryopreserved in heat-inactivated Human AB serum (GeminiBio) with 10% dimethylsulfoxide (Sigma-Aldrich) in liquid nitrogen. The serum was isolated and stored at −80 °C until further analyses.
Single-cell transcriptomics
Cryopreserved PBMCs were thawed and washed two times with PBS ×1. Cells were then counted using a Countess II Automated Cell Counter and loaded for scRNA-seq using ×10 Genomics Chromium Single Cell 30 Kit (v3.1 Chemistry). We preprocessed the reads with pyroe 0.9.0, Salmon 1.10.3 and alevin 0.10.0 packages with the reference genome GRCh38.p14 v46 from Gencode, and performed the downstream analysis with Seurat 5.1.0.
Longitudinal recipient samples were integrated with 22 external healthy control samples from 3 publicly available PBMC datasets (GEO accessions GSE165080, GSE171555 and GSE192391) using Harmony 1.2.0. The GSE165080 dataset consisted of 6 female samples and 5 male samples with a mean age of 43.1 ± 7.94 years39; the GSE171555 dataset included 3 female and 2 male samples40; and the GSE192391 dataset consisted of 4 female and 2 male samples41. Age information was not available for the GSE171555 and GSE192391 datasets.
Before integration, each dataset underwent independent quality control filtering followed by normalization using the Seurat SCTransform function. Based on an empirical evaluation of the distributions of the number of Unique Molecular Identifiers (UMIs) (nCount_RNA), number of detected genes (nFeature_RNA), log-transformed gene-to-UMI ratio (log10GenesPerUMI) and mitochondrial gene expression ratio (mitoRatio), the following filters were applied: GSE165080: nCount_RNA ≥ 1,800; nFeature_RNA ≥ 900 and GSE192391: nCount_RNA ≥ 1,800; nFeature_RNA ≥ 1,000 and GSE171555: nCount_RNA ≥ 1,800; nFeature_RNA ≥ 900 and
After integration, marker genes for each cluster were identified using the FindAllMarkers function following PrepSCTFindMarkers. Clusters were manually annotated into cell types based on the expression of the marker genes (Extended Data Fig. 3). A first round of annotation was performed at clustering resolutions of 0.2 and 0.4, resulting in 13 annotated cell types: CD4+ T cells, CD8+ T cells, erythrocytes, B cells, CD16+ monocytes, megakaryocytes/platelets, naive-like CD8+ T cells, plasmacytoid dendritic cells, conventional dendritic cells, two subsets of CD14+ monocytes and two subsets of NK cells. A second round of annotation was performed after sub-clustering aggregated monocyte and T cell subsets, followed by marker gene identification. This round annotated a third subset of CD14+ monocytes, naive-like CD4+ T cells, CD8+ MAIT cells and Treg cells. Single cells expressing canonical marker genes of at least two major cell types were assumed to be doublets and excluded from the Seurat object.
Gene sets of interest across time were identified empirically by calculating the average log2 fold change at each time point relative to the pretransplant baseline. This analysis was performed separately for CD14+ monocyte subsets, CD16+ monocytes and NK cell subsets. Genes were then ranked based on the absolute average log2 fold change to prioritize those with the most pronounced temporal shifts in expression.
Peripheral blood bulk transcriptomics
Venous blood collected at different time points and stored in PAXgene tubes at −80 °C were used for bulk RNA-seq. To process the bulk RNA-seq, we used FastQC 0.12.0 with standard parameters to evaluate the quality of the reads. We trimmed the adapters with Fastp 1.0.1 and quantified the abundance of transcripts with Salmon 1.10.3. We used the human reference transcriptome and the annotation named GRCh38.p14 v.46 from Gencode as the transcriptome reference. We used tximport 1.32.0 to import the abundance files and DESeq2 was used to harmonize samples. The same process was used for the public dataset (GSE120649 (ref. 27)). We used SRA tool 2.10.7 to download the public fastq files.
Proteomics
To perform proteomics analysis from serum samples, 11,000 markers were measured using SomaScan analysis (SomaLogic). This analysis was performed at the BIDMC Genomics, Proteomics, Bioinformatics and Systems Biology Center using the SomaScan Assay Kit for human serum, covering ~11,000 proteins. The expression counts were standardized using the median of hybridization controls. The outliers were flagged based on the limit of variation of 0.4–2.5 of the ratios.
Metabolomics
Untargeted metabolite analysis was performed using a Q Exactive HF-X mass spectrometer equipped with a HESI II probe and coupled to a Vanquish binary ultra-performance liquid chromatography system (Thermo Fisher Scientific). Each sample underwent two chromatographic separations. For both methods, 5 µl of the sample was injected into a BEH Z-HILIC column (100 × 2.1 mm, 1.7 µm, Waters). The first separation was conducted at pH 9, using 15 mM ammonium bicarbonate (Merck) in 90% water and 10% acetonitrile as mobile phase A, then 95% acetonitrile and 5% water as mobile phase B. The gradient was applied at a flow rate of 0.225 ml per min, following a method adapted from42, and the mass spectrometer operated in negative mode. The second separation followed the method of Mülleder et al.43, with mobile phases A and B buffered with 10 mM ammonium formate and 45 mM formic acid (pH 2.7). Mobile phase A consisted of a 1:1 acetonitrile to water mixture, while mobile phase B contained 95:5:5 acetonitrile to water to methanol. Chromatography was conducted at 40 °C (column oven) and 4 °C (autosampler), with a gradient flow rate of 0.4 ml per min as follows: an initial hold at 95% B for 0.75 min, a linear decrease to 30% B from 0.75 to 3.00 min, followed by a 1.00 min isocratic hold at 30% B. Mobile phase B was then returned to 95% over 0.50 min, with re-equilibration under initial conditions. Mass spectrometry was performed in positive mode. Data acquisition was carried out in full-scan mode, with the spray voltage set to 3 kV (negative mode) or 3.5 kV (positive mode). The capillary temperature was maintained at 320 °C, the HESI probe at 300 °C, the sheath gas at 40 U, the auxiliary gas at 8 U and the sweep gas at 1 U. The resolving power was set to 120,000. An untargeted metabolite library was generated using top-15 DDA acquisitions on a pooled study sample, with MS1 and MS2 resolutions of 60,000 and 30,000, respectively.
Raw data files (.raw) were processed using MZmine 4.544 and emzed45. Metabolite MS2 spectra were compared against HMDB, GNPS, MassBank and MoNA databases using spectral library matching (ref to DDA_library). In addition, retention times and m/z values from full-scan acquisitions of a pooled study sample were cross-referenced with an in-house database containing retention times of authenticated standards (Human Endogenous Metabolite Compound Library Plus L2501, TargetMol). Internal standards were integrated via emzed, and raw peak areas were normalized by dividing by sample biomass and the mean internal standard area.
Isolation of lymph node cells
On the day of the transplant, an iliac lymph node was harvested during the surgery for cell isolation. A lymph node from a patient who did not receive ATG was used as a control. The tissues were minced and digested with 500 U ml−1 collagenase D (Roche) for 30 min at 37 °C, followed by incubation with 0.1 M EDTA in PBS, pH 7.2, buffer for 5 min before a final suspension in 5 mM EDTA (Gibco), 1% FBS (Gibco) in × 1 PBS, pH 7.2 (Boston BioProducts). Isolated cells were then mechanically dissociated through a 70-μm cell strainer (Corning) in a 50-ml tube, and red blood cells were lysed using hypotonic ACK buffer (Gibco). Cells were then counted and cryopreserved in heat-inactivated human AB serum (GeminiBio) with 10% dimethylsulfoxide (Sigma-Aldrich) in liquid nitrogen.
Flow cytometry
Monitoring of immune cells over time was performed using 100 μl of whole blood. Cells were stained with surface antibodies for 30 min at room temperature, followed by incubation with 1× BD FACS Lysing Solution (BD Biosciences) for 10 min at room temperature for lysing red blood cells. We also stained cells isolated from human lymph nodes. Cells were thawed, washed twice and Fc-blocked (TruStain FcX, combination of anti-human CD16 (clone 3G8), CD32 (clone FUN-2) and CD64 (clone 10.1) antibodies, BioLegend, cat. no.: 422302) for 20 min before staining for surface markers for 30 min in FACS buffer (2% FBS in PBS × 1) on ice. The information about the antibodies used is shown in Supplementary Table 1. Stained cells were analyzed on an LSR Fortessa X-20 flow cytometer (BD Biosciences) with FACSDiva software (BD Biosciences) for all experiments. Data were analyzed using FlowJo software (Tree Star) in all experiments. Viable cells were selected based on the staining with LIVE/DEAD Fixable Blue Dead Cell Stain Kit (1:1000, Thermo Fisher, cat. no.: L23105) before Fc-blocking. Gating strategies for PBMC analysis were as previously described and can be found in Supplementary Fig. 2.
Innate immunity functional assay
Recipient’s PBMCs from different time points were thawed and washed twice with RPMI 1640 (Gibco) containing 10% FBS (GeminiBio) and ×1 pen/strep (Gibco) (complete RPMI), and then resuspended in complete RPMI. In a 96-well round-bottomed tissue culture plate at a volume of 100 μl, 4 × 105 cells per well were seeded. Cells were stimulated with 100 μl of either complete RPMI (controls) or a mixture of bacterial and viral TLR ligands (0.025 μg ml−1 LPS, 10 μg ml−1 Pam3CSK4, 4 μg ml−1 R848, 25 μg ml−1 poly I:C—all from Invivogen). PBMCs from each sample were stimulated in duplicates. After 24 h of incubation at 37 °C and 5% CO2, supernatants were obtained for Luminex analysis using the premixed ProcartaPlex Human Immune Monitoring Panel 65-plex (Thermo Fisher), including APRIL, BAFF, CXCL13, CD30, CD40L, CXCL5, CCL11, CCL24, CCL26, fibroblast growth factor-2 (FGF-2), CX3CL1, G-CSF, GM-CSF, CXCL1, hepatocyte growth factor, IFNα, IFNγ, IL-1α, IL-1β, IL-10, IL-12p70, IL-13, IL-15, IL-16, IL-17A, IL-18, IL-2, IL-20, IL-21, IL-22, IL-23, IL-27, IL-2R, IL-3, IL-31, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, CXCL10, CXCL11, LIF, CCL2, CCL7, CCL8, lymphotoxin-alpha, macrophage colony-stimulating factor, MDC, MIF, CXCL9, CCL3, CCL4, CCL20, MMP-1, nerve growth factor beta (NGF-β), SCF, SDF-1α, TNF, TNF-RII, TRAIL, TSLP, TWEAK and vascular endothelial growth factor A (VEGF-A). We read the assay in a Luminex200 machine, and data with a bead count
Intragraft tissue bulk transcriptomics
Bulk mRNA analysis was performed from formalin-fixed paraffin-embedded xenograft biopsies and a sample from the nontransplanted contralateral donor kidney, using the nCounter instrument (Nanostring) combined with the Banff Human Organ Transplant panel, following the methods and data analysis previously documented for human allograft biopsies25. The probe sequences were screened for homology with pig and NHP transcripts by a specialist at the manufacturer company (NanoString). We used probes from the panel that had >85% homology with porcine sequences for parenchymal cells or endothelium (n = 235 probes) and all probes for human leukocytes, similar to those used previously in pig-to-human xenografts5,30. Normalization was performed using seven housekeeping probes with >90% homology with the corresponding porcine transcripts. Pathways were manually curated from prior publications25,46,47,48 and KEGG Pathways (https://www.kegg.jp/kegg/pathway.html), Gene Ontology (geneontology.org) and Reactome (reactome.org) repositories. Cell types from tissue bulk were assessed with marker genes49.
Xenograft histopathologic analysis
Biopsy tissue was processed using standard kidney biopsy methods for light microscopy. hematoxylin and eosin samples were scored using the current Banff criteria50,51.
Multiplex immunofluorescence
Frozen slides from the donor collateral kidney biopsy and xenografts at days 8 and 34 were processed for multiplex imaging on the Orion platform (RareCyte), following Lin et al.52. An Orion multiplex antibody panel was used to profile immune cell subsets (Supplementary Table 2). In brief, slides were thawed at room temperature and then fixed in 4% paraformaldehyde in PBS for 45 min. They were washed in Tris‑buffered saline, permeabilized in 0.5% Triton X‑100 in Tris‑buffered saline for 5 min at room temperature, washed once in Tris‑buffered saline and once in PBS, and treated with 4.5% H2O2/24 mM NaOH in PBS (hydrogen peroxide solution) to reduce autofluorescence. After a surfactant wash and enhancer treatment, slides were incubated overnight at 4 °C with ArgoFluor‑conjugated antibodies and Hoechst 33342. The next day, slides were washed extensively in PBS, mounted in ArgoFluor mounting medium (RareCyte 42‑1214‑000), and imaged on an Orion microscope. Fluorophores were then quenched with hydrogen peroxide solution, and a second round of overnight 4 °C staining was performed with a different set of ArgoFluor‑conjugated antibodies (Supplementary Table 2) plus Hoechst 33342, followed by mounting and imaging.
Image stitching, segmentation and single‑cell quantification were carried out using the MCMICRO pipeline53. A Gaussian mixture model implemented in MATLAB (MathWorks)54 excluded cells exhibiting excessive autofluorescence. Gating strategies were defined as follows: CD4 T cells: HLA‑ABC+, CD45+ and CD4+; CD8 T cells: HLA‑ABC+, CD45+ and CD8+; M1 macrophages: HLA‑ABC+, CD68+ and CD163−; M2 macrophages: HLA‑ABC+ and CD163+; NK cells: HLA‑ABC+, CD45+ and NKG2A+. The HLA‑ABC marker recognizes human MHC I, thereby excluding from counts the few residual donor pig kidney-resident M2 macrophages with which the CD163 antibody cross‑reacts (Extended Data Fig. 7).
DSA quantification using IgG binding assay
After pig donor blood collection, PBMCs were isolated using Lympholyte-Mammal Cell Separation Media (Cedarlane) according to the manufacturer’s protocol. Using a V-bottom 96-well-plate, serum was serially diluted in PBS ×1 as follows: 1:4, 1:8, 1:16, 1:32, 1:64 and 1:128. Serum from a healthy human control and a Baboon previously transplanted with a pig kidney were used as negative and positive controls, respectively, along with different time points of the recipient serum. To avoid nonspecific binding sites, pig PBMCs (5 × 105 cells per well) were blocked with 10% heat-inactivated goat serum (R&D Systems, cat. no.: S13110H) for 20 mins at 4 °C. After two washes, pig PBMCs were incubated with serially diluted serum for 1 h at 4 °C and washed three times in cold PBS × 1. Next, pig PBMCs were incubated for 1 h at 4 °C with anti-human IgG Fc-APC (clone M1310G05, BioLegend). After two washes, cells were resuspended in a staining buffer (PBS, 1% of bovine serum albumin, 0.09% sodium azide). Stained cells were analyzed on an LSR Fortessa X-20 flow cytometer (BD Biosciences) with FACSDiva software (BD Biosciences) for all experiments. Data were analyzed using FlowJo software (Tree Star) in all experiments.
Cell-free DNA quantification
Karius retrieved porcine and human cfDNA using a protocol similar to that described by Blauwkamp et al.55. Briefly, plasma samples were initially processed by centrifugation to remove residual cells, after which cfDNA was extracted using a modified Mag-Bind cfDNA kit in automated liquid handling platforms. Sequencing libraries were prepared with dual-indexed Ovation Ultralow System kits and sequenced on Illumina NextSeq sequencers. The resulting reads underwent stringent bioinformatics processing, including alignment against human, porcine and microbial reference databases to distinguish human and porcine cfDNA fragments accurately. The concentration of each species’ cfDNA was quantitatively estimated by normalizing against synthetic internal control molecules (WINC molecules), ensuring precise measurement of cfDNA abundance in plasma samples
Functional data analysis
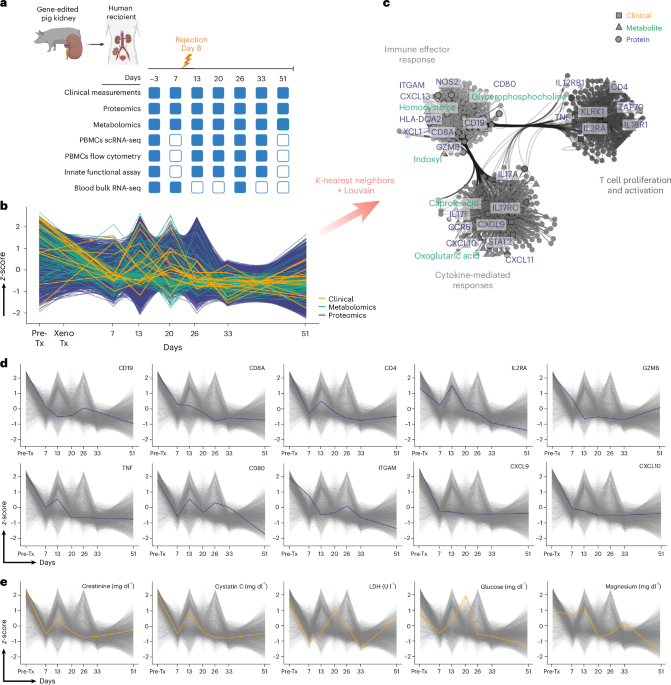
We normalized all the data series to z-score with scikit-learn 1.6.156. To analyze the data in time, we used scikit-fda 0.9.157. With this package, we excluded outliers using the MS-Plot Outlier Detector method. We then used Fuzzy C-Means with l2-distance to cluster. We defined the optimal number of three clusters, calculating the silhouette score with scikit-learn 1.6.1. To plot the time series, we calculated the median with NumPy 2.2.0 and plotted the line and the violins with Matplotlib 3.8.0.
Network analysis
For the three clusters defined by functional data analysis, we used the z-score normalized proteomics data to calculate the nearest neighbors with scikit-fda 0.9.1, calculated the Louvain communities with NetworkX 3.4.258 based on the connectivity of the nearest neighbor’s graph, followed by an enrichment analysis for each community with GSEApy 1.1.759. After annotating the clusters based on GSEApy results, we trained a k-neighbors classifier with scikit-fda 0.9.1 on proteomics Louvain labels. We predicted the Louvain communities for z-score normalized metabolomics data. To plot networks, we used Netgraph 4.13.1 to annotate the nodes. We used the package adjustText 1.3.0 (https://github.com/Phlya/adjustText).
Cell population abundance—Cibersortx
After preprocessing the whole-blood bulk transcriptomic data, we normalized the expression counts with DESeq2. Next, we used the R package IOBR 0.99.860 to estimate the population abundance on whole blood using the LM22 and LM6 matrices without tumor optimization.
Intersection of differentially expressed proteins and genes
We calculated the log2 fold change between days 7 and 26 (log2[D7/D26]) for both transcriptomics and proteomics. Since we have only one sample, we selected the most relevant fold change by the first quartile of negative fold changes and the third quartile of positive fold changes to define downregulated and upregulated molecules, respectively. We found the intersection using Python native functions and performed an enrichment analysis with GSEApy 1.1.759. To plot the Venn diagram, we used the venn 0.1.3 package.
Chord diagram visualization
A chord diagram was constructed using the circlize package 0.4.1661 in R 4.4.2 to illustrate relationships between differentially expressed genes and proteins. Sectors representing upregulated and downregulated genes and proteins were color coded, and interconnections were drawn based on shared elements between sets. Edge transparency and arrow annotations were applied to improve interpretability.
Comparative analysis of xenograft and allograft rejection profiles
To maintain consistency, we reprocessed the public dataset GSE120649 (ref. 27) from whole-blood human kidney transplant recipients using the same pipeline used for the xenotransplant samples. We used a previously described transcriptional profile of human allograft rejection28 to compare allo and xenotransplantation gene expression. To detect nonlinear correlation, we clustered the samples using the complete linkage method on the distance correlation metric with SciPy 1.11.462. We used seaborn 0.13.263 to plot the cluster heatmap.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.